Cython and Numba applied to a simple algorithm
The aim of this notebook is to show a basic example of Cython and Numba, applied to a simple algorithm: Insertion sort.
As we will see, the code transformation from Python to Cython or Python to Numba can be really easy (specifically for the latter), and results in very efficient code for sorting algorithms. This is due to the fact that the computer is CPU bound when executing this type of algorithmic task, for which the overhead of calling the CPython API in pure Python is really large.
Let us recall the purpose of these two Python-related tools from their respective websites:
Cython is an optimizing static compiler for both the Python programming language and the extended Cython programming language
Numba is an open source JIT compiler that translates a subset of Python and NumPy code into fast machine code.
Now, let’s describe the chosen algorithm: Insertion sort, which is a very simple and intuitive algorithm. As written in Cormen et al. [1]:
Insertion sort works the way many people sort a hand of playing cards. We start with an empty left hand and the cards face down on the table. We then remove one card at a time from the table and insert it into the correct position in the left hand. To find the correct position for a card, we compare it with each of the cards already in the hand, from right to left […]. At all times, the cards held in the left hand are sorted, and these cards were originally the top cards of the pile on the table.
Here is a visualization of the Insertion sort process applied to 25 random elements (the code used to generate this animated gif is shown at the end of the notebook):
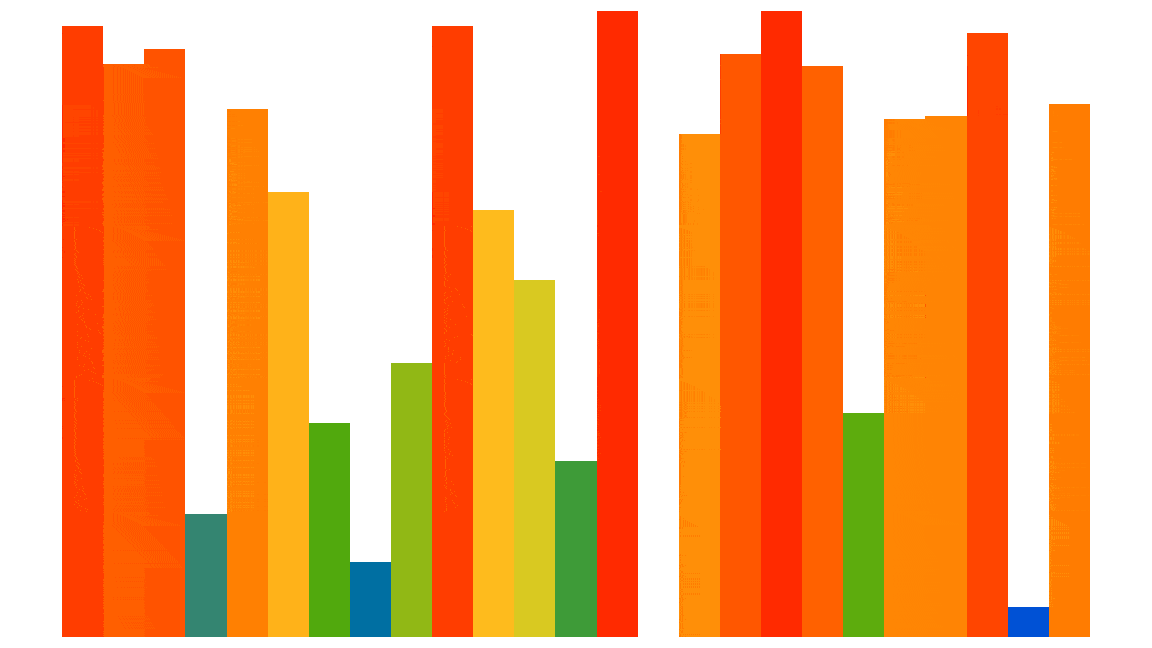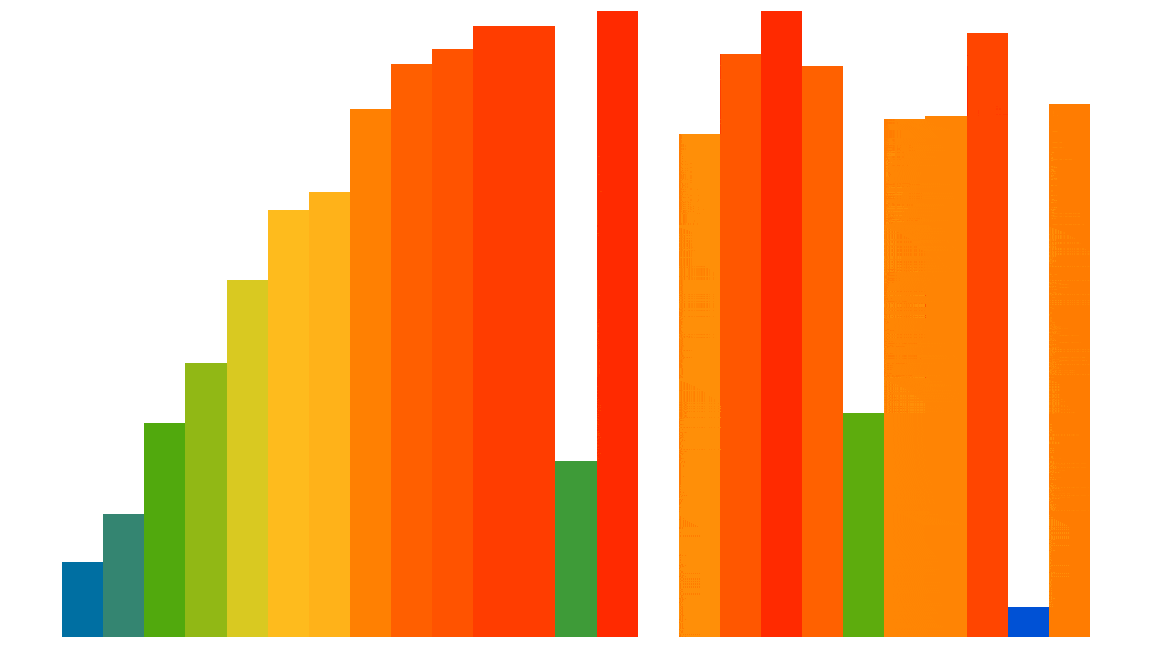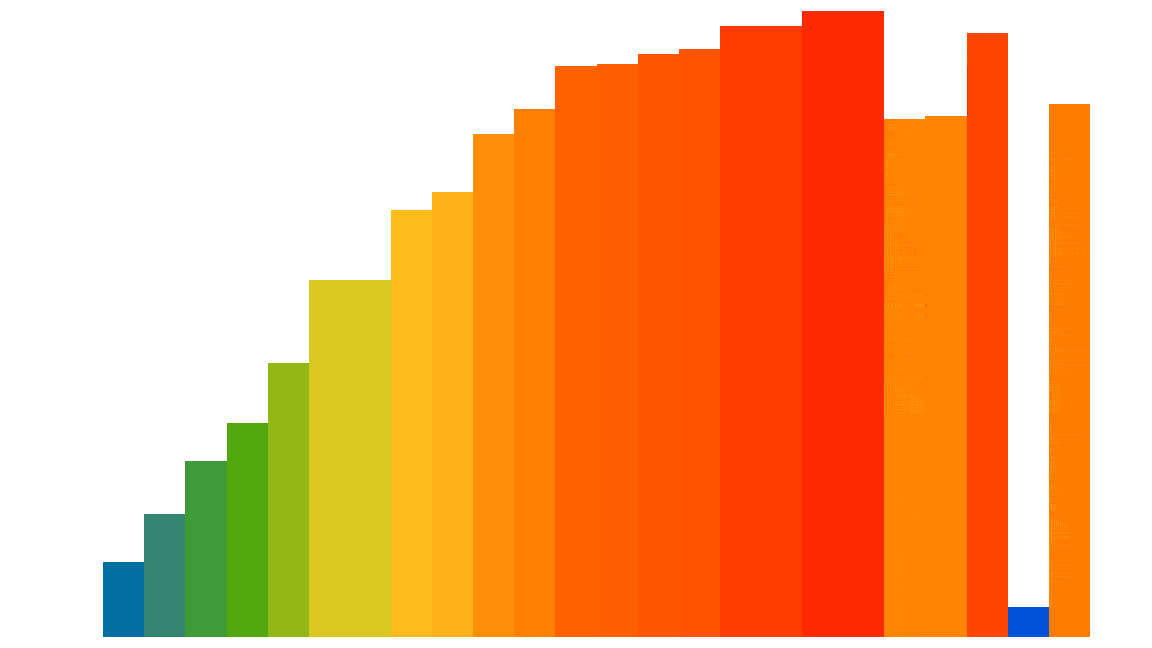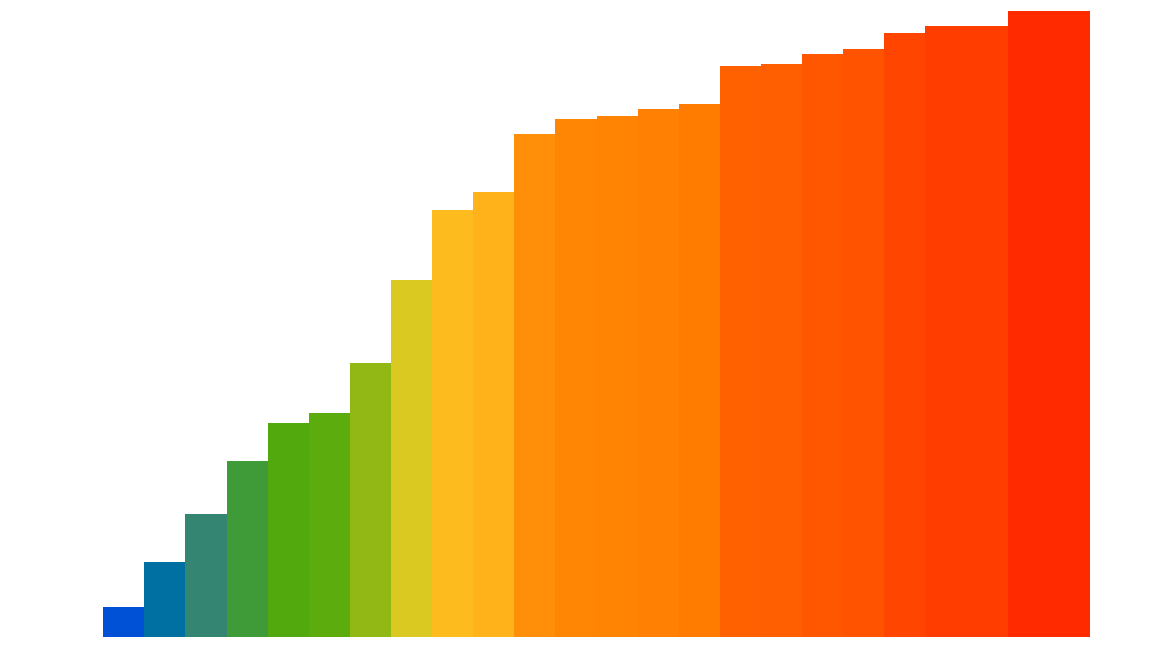
However, this algorithm is not so efficient, except for elements that are almost already sorted: its performance is quadratic, i.e. $О ( n^2 )$. But we are only interested here in comparing different optimization approaches in Python and not actually in sorting efficiently.
Here is the Python code for an in-place array-based implementation:
for j in range(1, len(A)):
key = A[j]
i = j - 1
while (i >= 0) & (A[i] > key):
A[i + 1] = A[i]
i = i - 1
A[i + 1] = key
And here are some other facts about Insertion sort from Wikipedia:
- Adaptive, i.e., efficient for data sets that are already substantially sorted: the time complexity is $ O(kn) $ when each element in the input is no more than $k$ places away from its sorted position
- Stable, i.e., does not change the relative order of elements with equal keys
- In-place, i.e., only requires a constant amount $ O(1) $ of additional memory space
- Online, i.e., can sort a list as it receives it
[1] Introduction to Algorithms, T. Cormen, C. Leiserson, R. Rivest, and C. Stein. The MIT Press, 3rd edition, (2009)
Now here is the code.
Imports
perfplot is used to measure runtime for all different combination of array length and method.
import itertools
import numpy as np
import matplotlib.pyplot as plt
import perfplot
from numba import jit
%load_ext Cython
np.random.seed(124) # Seed the random number generator
Python implementation
def insertion_sort_inplace_python(A):
for j in range(1, len(A)):
key = A[j]
i = j - 1
while (i >= 0) & (A[i] > key):
A[i + 1] = A[i]
i = i - 1
A[i + 1] = key
Numba implementation
As you can observe, this is stricly the same as the pure Python implementation, except for the @jit (just-in-time) decorator:
@jit(nopython=True)
def insertion_sort_inplace_numba(A):
for j in range(1, len(A)):
key = A[j]
i = j - 1
while (i >= 0) & (A[i] > key):
A[i + 1] = A[i]
i = i - 1
A[i + 1] = key
Cython implementation
Again, this is very similar to the Python implementation, especially the looping part.
%%cython
import cython
cimport numpy as cnp
@cython.boundscheck(False)
@cython.wraparound(False)
@cython.initializedcheck(False)
cpdef void insertion_sort_inplace_cython_int64(cnp.int64_t[:] A) nogil:
cdef:
Py_ssize_t i, j
cnp.int64_t key
int length_A = A.shape[0]
for j in range(1, length_A):
key = A[j]
i = j - 1
while (i >= 0) & (A[i] > key):
A[i + 1] = A[i]
i = i - 1
A[i + 1] = key
The differences are the following ones:
- we add the
%%cythonmagic for interactive work with Cython in Jupyterlab - we import some libraries (
cythonand the NumPy C API) specifically for thic Cython notebook cell - we add some compiler directives (instructions which affect which kind of code Cython generates). Here is a decription of the various compiler directives from the Cython documentation
- the function is defined as
cpdefwhich means that it can be called either from some Python or Cython code. In our case, we are going to call it from a Python function - in the arguments, a 1D typed memoryview is performed on the given NumPy
int64array:cnp.int64_t[:] A, which allows a fast/direct access to memory buffers. However, since this is typed, we need to write another function if dealing with floats, e.g. with acnp.float64_t[:]memoryview. - all variables within the function are declared
nogilis added at the end of the function signature, to indicate the release of the GIL. In the present case, this is only to make sure that the CPython API is not used within the function (or there would be an error when executing the cell).
Main function
def insertion_sort(A, kind):
B = np.copy(A)
if kind == 'python':
insertion_sort_inplace_python(B)
elif kind == 'cython':
insertion_sort_inplace_cython_int64(B)
elif kind == 'numba':
insertion_sort_inplace_numba(B)
return B
Timings
First, we check that the result is invariant with respect to the function called:
N = 100
A = np.random.randint(low=0, high=10 * N, size=N, dtype=np.int64)
A_sorted = np.sort(A)
A_sorted_cython = insertion_sort(A, kind='cython')
A_sorted_python = insertion_sort(A, kind='python')
A_sorted_numba = insertion_sort(A, kind='numba')
np.testing.assert_array_equal(A_sorted_cython, A_sorted_python)
np.testing.assert_array_equal(A_sorted_cython, A_sorted_numba)
np.testing.assert_array_equal(A_sorted_cython, A_sorted)
Then we compare the execution time of the four different implementations: Python, Cython, Numba and NumPy. The NumPy command is np.sort with the default quicksort algorithm (implemented in C).
With pure Python
out = perfplot.bench(
setup=lambda n: np.random.randint(low=0, high=10 * n, size=n, dtype=np.int64),
kernels=[
lambda A: insertion_sort(A, kind='python'),
lambda A: insertion_sort(A, kind='cython'),
lambda A: insertion_sort(A, kind='numba'),
lambda A: np.sort(A),
],
labels=['Python', 'Cython', 'Numba', 'NumPy'],
n_range=[10**k for k in range(1, 4)],
)
ms = 10
fig = plt.figure(figsize=(8, 8))
ax = fig.add_subplot(1, 1, 1)
plt.loglog(out.n_range, np.power(out.n_range, 2) * 1.e-9, 'o-', label='$c \; n^2$')
plt.loglog(out.n_range, out.timings[1] * 1.e-9, 'o-', ms=ms, label='Cython')
plt.loglog(out.n_range, out.timings[2] * 1.e-9, 'o-', ms=ms, label='Numba')
plt.loglog(out.n_range, out.timings[3] * 1.e-9, 'o-', ms=ms, label='NumPy')
plt.loglog(out.n_range, out.timings[0] * 1.e-9, 'o-', ms=ms, label='Python')
markers = itertools.cycle(("", "o", "v", "^", "<", ">", "s", "p", "P", "*", "h", "X", "D", '.'))
for i, line in enumerate(ax.get_lines()):
marker = next(markers)
line.set_marker(marker)
plt.legend()
plt.grid('on')
_ = ax.set_ylabel('Runtime [s]')
_ = ax.set_xlabel('n = len(A)')
_ = ax.set_title('Timings of Insertion sort')
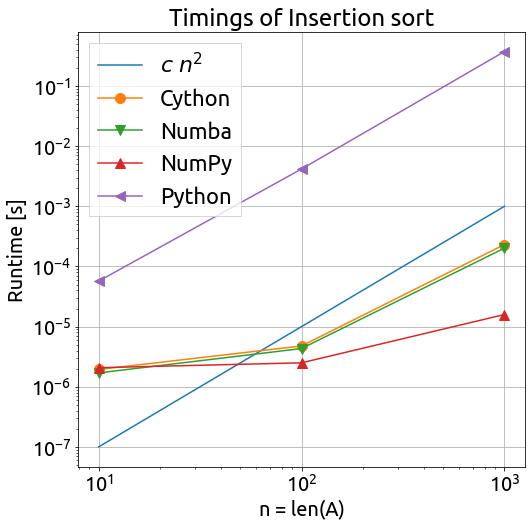
We can observe the following things regarding the execution time:
- pure Python is slower by a factor 100 to 1000
- the Cython and Numba implementations are very close, and probably equivalent to C
- the quicksort NumPy algorithm is way more efficient ($O(n \; log \; n)$ on average)
Without pure Python
Let us run again the comparison without the pure Python version this time, in order to sort larger arrays.
out = perfplot.bench(
setup=lambda n: np.random.randint(low=0, high=10 * n, size=n, dtype=np.int64),
kernels=[
lambda A: insertion_sort(A, kind='cython'),
lambda A: insertion_sort(A, kind='numba'),
lambda A: np.sort(A),
],
labels=['Cython', 'Numba', 'NumPy'],
n_range=[10**k for k in range(1, 6)],
)
fig = plt.figure(figsize=(8, 8))
ax = fig.add_subplot(1, 1, 1)
plt.loglog(out.n_range, np.power(out.n_range, 2) * 1.e-9, 'o-', label='$c \; n^2$')
plt.loglog(out.n_range, out.timings[0] * 1.e-9, 'o-', ms=ms, label='Cython')
plt.loglog(out.n_range, out.timings[1] * 1.e-9, 'o-', ms=ms, label='Numba')
plt.loglog(out.n_range, out.timings[2] * 1.e-9, 'o-', ms=ms, label='NumPy')
markers = itertools.cycle(("", "o", "v", "^", "<", ">", "s", "p", "P", "*", "h", "X", "D", '.'))
for i, line in enumerate(ax.get_lines()):
marker = next(markers)
line.set_marker(marker)
plt.legend()
plt.grid('on')
_ = ax.set_ylabel('Runtime [s]')
_ = ax.set_xlabel('n = len(A)')
_ = ax.set_title('Timings of Insertion sort')
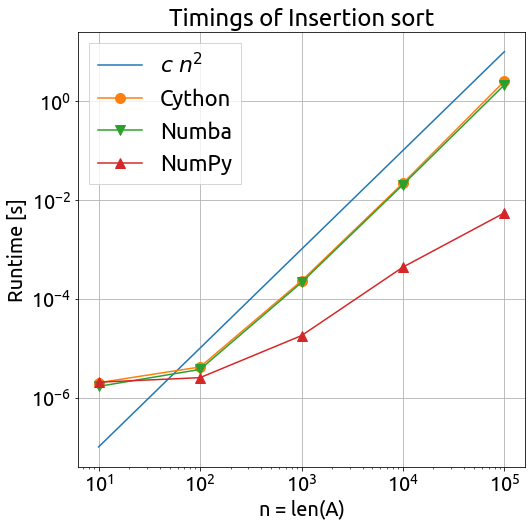
Conclusion
We can see that both Cython and Numba give very good results regarding the optimization of NumPy-based Python code. Numba is easier to use but I think that Cython is more flexible regarding the kinds of algorithms that you can optimize, although a little bit more complex.
Appendix: generate the animated gif
This is done by dumping many matplotlib png figures into a folder and then aggregating the images into an animation using the convert linux command.
from matplotlib.colors import Normalize
from colorcet import palette
import matplotlib as mpl
def plot(A, k, high, cmap):
norm = Normalize(vmin=0, vmax=1)
fig = plt.figure(figsize=(16, 9))
ax = fig.add_subplot(1, 1, 1)
_ = plt.bar(np.arange(len(A)), height=A, color=cmap(norm(A / high)), width=1.0)
_ = ax.set_ylim(0, high)
_ = plt.axis('off')
fig.tight_layout()
_ = plt.savefig(f'./images/Insertion_sort_{str(k).zfill(3)}.png', color=cmap(norm(A / high)))
plt.close()
def insertion_sort_inplace_python(A, cmap):
high = np.max(A)
k = 0
plot(A, k, high, cmap)
k += 1
for j in range(1, len(A)):
key = A[j]
i = j - 1
while (i >= 0) & (A[i] > key):
A[i + 1] = A[i]
i = i - 1
plot(A, k, high, cmap)
k += 1
A[i + 1] = key
plot(A, k, high, cmap)
k += 1
if False:
colorcet_cmap = 'rainbow'
cmap = mpl.colors.ListedColormap(palette[colorcet_cmap], name=colorcet_cmap) # register the colorcet colormap
!mkdir -p ./images/
N = 25
A = np.random.randint(low=0, high=10 * N, size=N, dtype=np.int64)
!rm ./images/*.png
insertion_sort_inplace_python(A, cmap)
!convert -delay 1 -loop 0 ./images/Insertion_sort_*.png animation.gif