Optuna + XGBoost on a tabular dataset
Updated Sep 16, 2021 following a comment by @k_nzw about XGBoostPruningCallback
The purpose of this Python notebook is to give a simple example of hyperparameter optimization (HPO) using Optuna and XGBoost. We are going to perform a regression on tabular data with single output.
XGBoost is a well-known gradient boosting library, with some hyperparameters, and Optuna is a powerful hyperparameter optimization framework. Tabular data still are the most common type of data found in a typical business environment.
We are going to use a dataset from Kaggle : Tabular Playground Series - Feb 2021. These playground competitions are great for practicing machine learning skills. If you have a kaggle account and installed the kaggle package, you can download the data by running :
kaggle competitions download -c tabular-playground-series-feb-2021
Note that this is not exactly real-world data. As described in the competition page :
The dataset used for this competition is synthetic, but based on a real dataset and generated using a CTGAN. The original dataset deals with predicting the amount of an insurance claim. Although the features are anonymized, they have properties relating to real-world features.
An important point is that we are not going to perform an Exploratory Data Analysis (EDA) or any Feature Engineering (FE) besides what is stricly necessary in order to use XGBoost. The only focus of this post is hyperparameter optimization of XGBoost with Optuna and it would be too long to describe here the whole process of making a model with a new dataset.
Imports
import os
import string
import numpy as np
import pandas as pd
from sklearn.model_selection import RepeatedKFold
from sklearn.metrics import mean_squared_error
from sklearn.ensemble import RandomForestRegressor
from sklearn.experimental import enable_hist_gradient_boosting
from sklearn.ensemble import HistGradientBoostingRegressor
from xgboost import XGBRegressor
from optuna import create_study
from optuna.samplers import TPESampler
from optuna.integration import XGBoostPruningCallback
FS = (14, 6) # figure size
RS = 124 # random state
N_JOBS = 8 # number of parallel threads
# repeated K-folds
N_SPLITS = 10
N_REPEATS = 1
# Optuna
N_TRIALS = 100
MULTIVARIATE = True
# XGBoost
EARLY_STOPPING_ROUNDS = 100
The package versions are the following ones :
Python : 3.8.6
pandas : 1.2.1
xgboost : 1.3.0
sklearn : 0.24.1
optuna : 2.5.0
numpy : 1.19.5
Loading the data
train_df = pd.read_csv("./train.csv", index_col=0)
test_df = pd.read_csv("./test.csv", index_col=0)
Quick data preparation
Let’s have a look at this dataset :
train_df.shape
(300000, 25)
train_df.head(3)
| cat0 | cat1 | ... | cont13 | target | |
|---|---|---|---|---|---|
| id | |||||
| 1 | A | B | ... | 0.719903 | 6.994023 |
| 2 | B | A | ... | 0.808464 | 8.071256 |
| 3 | A | A | ... | 0.828352 | 5.760456 |
3 rows × 25 columns
We have 24 feature and 1 target columns (10 categorical and 14 continuous features) :
train_df.dtypes
cat0 object
cat1 object
cat2 object
cat3 object
cat4 object
cat5 object
cat6 object
cat7 object
cat8 object
cat9 object
cont0 float64
cont1 float64
cont2 float64
cont3 float64
cont4 float64
cont5 float64
cont6 float64
cont7 float64
cont8 float64
cont9 float64
cont10 float64
cont11 float64
cont12 float64
cont13 float64
target float64
dtype: object
cols = train_df.columns
cat_cols = [c for c in cols if c.startswith("cat")] # categorical features
cont_cols = [c for c in cols if c.startswith("cont")] # continuous features
feature_cols = cat_cols + cont_cols
target_col = "target"
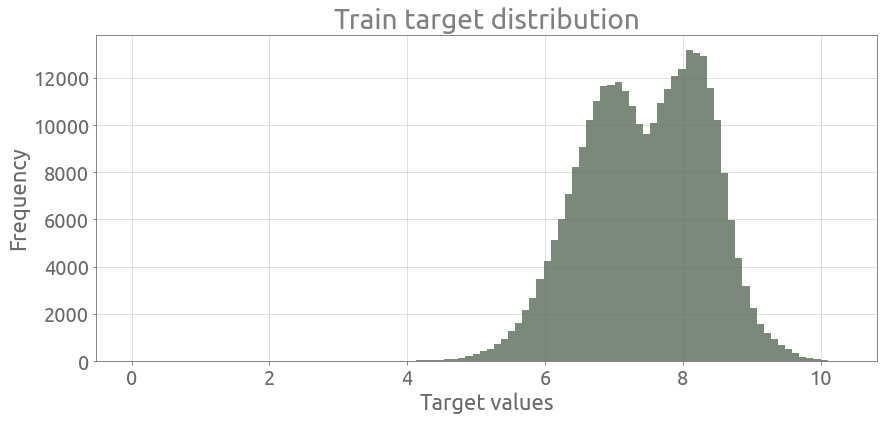
Here is the distribution of the target :
ax = train_df.target.plot.hist(bins=100, figsize=FS, alpha=0.6)
ax.grid()
_ = ax.set(title="Train target distribution", xlabel="Target values")
There is no missing data (not a very common situation!) :
train_df.isna().any().any()
False
test_df.isna().any().any()
False
Categorical feature encoding
We need to transform the categorical features into numerical values. Let’s see the number of distinct values in each categorical feature :
train_df[cat_cols].nunique()
cat0 2
cat1 2
cat2 2
cat3 4
cat4 4
cat5 4
cat6 8
cat7 8
cat8 7
cat9 15
dtype: int64
We can also display the distinct values in each categorical feature along with the respective value count :
for i, cat_col in enumerate(cat_cols):
cat_col = "cat" + str(i)
print(
f"{cat_col} :",
dict(
zip(
list(np.sort(train_df[cat_col].unique())),
list(train_df[cat_col].value_counts().values),
)
),
)
cat0 : {'A': 281471, 'B': 18529}
cat1 : {'A': 162678, 'B': 137322}
cat2 : {'A': 276551, 'B': 23449}
cat3 : {'A': 183752, 'B': 104464, 'C': 11174, 'D': 610}
cat4 : {'A': 297373, 'B': 1241, 'C': 767, 'D': 619}
cat5 : {'A': 149208, 'B': 135151, 'C': 11763, 'D': 3878}
cat6 : {'A': 292643, 'B': 6344, 'C': 809, 'D': 147, 'E': 24, 'G': 19, 'H': 11, 'I': 3}
cat7 : {'A': 267631, 'B': 24356, 'C': 5750, 'D': 1961, 'E': 279, 'F': 14, 'G': 6, 'I': 3}
cat8 : {'A': 121054, 'B': 94616, 'C': 42195, 'D': 37878, 'E': 3694, 'F': 549, 'G': 14}
cat9 : {'A': 107281, 'B': 50064, 'C': 42200, 'D': 24759, 'E': 20955, 'F': 13408, 'G': 10409, 'H': 9838, 'I': 6981, 'J': 6173, 'K': 4112, 'L': 3435, 'M': 209, 'N': 103, 'O': 73}
We are going to use some basic ordinal encoding :
alphabet = string.ascii_uppercase
mapping = dict(zip(alphabet, range(len(alphabet))))
train_df[cat_cols] = train_df[cat_cols].replace(mapping)
test_df[cat_cols] = test_df[cat_cols].replace(mapping)
train_df.head(3)
| cat0 | cat1 | ... | cont13 | target | |
|---|---|---|---|---|---|
| id | |||||
| 1 | 0 | 1 | ... | 0.719903 | 6.994023 |
| 2 | 1 | 0 | ... | 0.808464 | 8.071256 |
| 3 | 0 | 0 | ... | 0.828352 | 5.760456 |
3 rows × 25 columns
We are now ready to use XGBoost :
X_train = train_df[feature_cols]
X_test = test_df[feature_cols]
y_train = train_df[target_col]
Baseline
Here is a little function used to evaluate a given model object that has a scikit-learn interface (.fit(), .predict() methods) :
def evaluate_model_rkf(model, X_df, y_df, n_splits=5, n_repeats=2, random_state=63):
X_values = X_df.values
y_values = y_df.values
rkf = RepeatedKFold(
n_splits=n_splits, n_repeats=n_repeats, random_state=random_state
)
y_pred = np.zeros_like(y_values)
for train_index, test_index in rkf.split(X_values):
X_A, X_B = X_values[train_index, :], X_values[test_index, :]
y_A = y_values[train_index]
model.fit(
X_A, y_A,
)
y_pred[test_index] += model.predict(X_B)
y_pred /= n_repeats
return np.sqrt(mean_squared_error(y_train, y_pred))
We use a repeated k-fold cross-validation for model evaluation. Actually, because the dataset is sufficiently large (300000 samples), we do not repeat the k-fold process in the following (n_repeats=1). The collection of all the out-of-fold predictions are being used to compute the model performance, Root Mean Square Error (RMSE), of the full training dataset.
Let’s try some models from scikit-learn, such as RandomForestRegressor and HistGradientBoostingRegressor with default settings :
model = RandomForestRegressor(random_state=RS, n_jobs=N_JOBS)
evaluate_model_rkf(
model, X_train, y_train, n_splits=N_SPLITS, n_repeats=N_REPEATS, random_state=RS
)
0.8589072366431951
model = HistGradientBoostingRegressor(random_state=RS)
evaluate_model_rkf(
model, X_train, y_train, n_splits=N_SPLITS, n_repeats=N_REPEATS, random_state=RS
)
0.8465656534244064
Note that HistGradientBoostingRegressor uses all the default cores by default. We can also evaluate a XGBoost model with default settings :
model = XGBRegressor(seed=RS, n_jobs=N_JOBS)
evaluate_model_rkf(
model, X_train, y_train, n_splits=N_SPLITS, n_repeats=N_REPEATS, random_state=RS
)
0.8485711113354382
If we have a look at the leaderboard of the competition, we can see that the best RMSE scores are between 0.841 and 0.842 at the the time of writing this post. The 3 algorithms above with default settings leads to scores above 0.846, with HistGradientBoostingRegressorbeing by far the most efficient if we also take computational time into account. Anyway, let’s try to tune the parameters of XGBoost in order to decrease this score.
Optuna + XGBoost
Let’s define an objective function for the optimization process. With Optuna, a Trial instance represents a process of evaluating an objective function with various suggested values. Optuna can suggest different kind of parameters :
suggest_categoricalsuggest_loguniformsuggest_intsuggest_discrete_uniformsuggest_floatsuggest_uniform
Even if Optuna is a great library, we should try to make the optimization problem easier by reducing the search space. XGBoost has at least a dozen of hyperparameters. We are using here the Scikit-Learn API of XGBoost. Here is a list of some parameters of this interface :
n_estimators(int) – Number of gradient boosted trees.max_depth(int) – Maximum tree depth for base learners.learning_rate(float) – Boosting learning rate.booster(string) – Specify which booster to use: gbtree, gblinear or dart.tree_method(string) – Specify which tree method to use.gamma(float) – Minimum loss reduction required to make a further partition on a leaf node of the tree.min_child_weight(float) – Minimum sum of instance weight(hessian) needed in a child.max_delta_step(float) – Maximum delta step we allow each tree’s weight estimation to be.subsample(float) – Subsample ratio of the training instance.colsample_bytree(float) – Subsample ratio of columns when constructing each tree.colsample_bylevel(float) – Subsample ratio of columns for each level.colsample_bynode(float) – Subsample ratio of columns for each split.reg_alpha(float) – L1 regularization term on weightsreg_lambda(float) – L2 regularization term on weights
In this post, we are not going into much details about the gradient boosting algorithm and all the different parameters.
A pragmatic approach is to use a large number of n_estimators and then activates early stopping with early_stopping_rounds (we use early_stopping_rounds=100 in this post) in the fit()method :
Validation metric needs to improve at least once in every
early_stopping_roundsround(s) to continue training.
Then, some of the most important parameters are learning_rate, max_depth, min_child_weight. In maybe a little lower level of importance comes the parameters subsample, colsample_bytree and the regularization terms.
So we can imagine to start by tuning the learning_rate and then adjust sequentially some groups of parameters, by order of importance. But here we are going to optimize most of these parameters all together, to make it shorter.
Also, an important setting is the interval range for each parameter. That would be kind of very optimistic to set very wide search intervals for each parameter, so we are going to reduce these intervals. This is a really empirical process for me here and I actually looked at other kaggle kernels using XGBoost to limit the search space (this very interesting kernel by Bojan Tunguz for example).
Remarks :
- Unpromising trials are pruned using
XGBoostPruningCallback, based on the RMSE on the current validation fold. - We set n_jobs=8 (the number of cores of my laptop) for XGBoost and 1 for the HPO process.
Update : Following an insightful comment by @k_nzw, I understood that it is not appropriate to use the pruning callback within k-fold cross validation. It appears that it is meant to be used when there is a single training per trial. As explained by @k_nzw:
This is because at each trial, we can report intermediate value once at each step.
So in our case with several trainings per trial, the callback might only be used in the first step of the cross validation loop but not in the following steps… Which is not what I expected. Thanks again @k_nzw for your comment, ありがとう!Although the pruning is kind of useless here, I keep the code as it was first written and hope that someone else might learn from this mistake.
So here is the objective function :
def objective(
trial,
X,
y,
random_state=22,
n_splits=3,
n_repeats=2,
n_jobs=1,
early_stopping_rounds=50,
):
# XGBoost parameters
params = {
"verbosity": 0, # 0 (silent) - 3 (debug)
"objective": "reg:squarederror",
"n_estimators": 10000,
"max_depth": trial.suggest_int("max_depth", 4, 12),
"learning_rate": trial.suggest_loguniform("learning_rate", 0.005, 0.05),
"colsample_bytree": trial.suggest_loguniform("colsample_bytree", 0.2, 0.6),
"subsample": trial.suggest_loguniform("subsample", 0.4, 0.8),
"alpha": trial.suggest_loguniform("alpha", 0.01, 10.0),
"lambda": trial.suggest_loguniform("lambda", 1e-8, 10.0),
"gamma": trial.suggest_loguniform("lambda", 1e-8, 10.0),
"min_child_weight": trial.suggest_loguniform("min_child_weight", 10, 1000),
"seed": random_state,
"n_jobs": n_jobs,
}
model = XGBRegressor(**params)
pruning_callback = XGBoostPruningCallback(trial, "validation_0-rmse")
rkf = RepeatedKFold(
n_splits=n_splits, n_repeats=n_repeats, random_state=random_state
)
X_values = X.values
y_values = y.values
y_pred = np.zeros_like(y_values)
for train_index, test_index in rkf.split(X_values):
X_A, X_B = X_values[train_index, :], X_values[test_index, :]
y_A, y_B = y_values[train_index], y_values[test_index]
model.fit(
X_A,
y_A,
eval_set=[(X_B, y_B)],
eval_metric="rmse",
verbose=0,
callbacks=[pruning_callback],
early_stopping_rounds=early_stopping_rounds,
)
y_pred[test_index] += model.predict(X_B)
y_pred /= n_repeats
return np.sqrt(mean_squared_error(y_train, y_pred))
Now let’s define a sampler. Optuna provides a Tree-structured Parzen Estimator (TPE) algorithm with TPESampler . We also need to create a study with create_study in order to start the optimization process. Here is a paper with some references about the algorithms found in Optuna. n_trials is the number of objective evaluations, set to 100 in the following.
Note that we activate the experimantal multivariate option of the TPE sampler.
sampler = TPESampler(seed=RS, multivariate=MULTIVARIATE)
study = create_study(direction="minimize", sampler=sampler)
study.optimize(
lambda trial: objective(
trial,
X_train,
y_train,
random_state=RS,
n_splits=N_SPLITS,
n_repeats=N_REPEATS,
n_jobs=8,
early_stopping_rounds=EARLY_STOPPING_ROUNDS,
),
n_trials=N_TRIALS,
n_jobs=1,
)
# display params
hp = study.best_params
for key, value in hp.items():
print(f"{key:>20s} : {value}")
print(f"{'best objective value':>20s} : {study.best_value}")
We do not display the log here, which is kind of verbose. Here are the final parameter values found by Optuna, and the corresponding objective value :
max_depth : 8
learning_rate : 0.037288466802750865
colsample_bytree : 0.3301265198894751
subsample : 0.598344890923238
alpha : 0.01320580211991565
lambda : 7.527644719697382e-08
min_child_weight : 837.0649573787646
best objective value : 0.8425081635928959
So we should get a score between 0.842 and 0.843 on the test set.
Submit and evaluate the prediction
We are going to retrain the model with the optimal parameter dictionary hp, make a prediction on the test dataset and submit this prediction on the kaggle website :
hp["verbosity"] = 0
hp["objective"] = "reg:squarederror"
hp["n_estimators"] = 10000
hp["seed"] = RS
hp["n_jobs"] = 8
model = XGBRegressor(**hp)
rkf = RepeatedKFold(n_splits=N_SPLITS, n_repeats=N_REPEATS, random_state=RS)
X_values = X_train.values
y_values = y_train.values
y_pred = np.zeros_like(test_df.cont0.values)
for train_index, test_index in rkf.split(X_values):
X_A, X_B = X_values[train_index, :], X_values[test_index, :]
y_A, y_B = y_values[train_index], y_values[test_index]
model.fit(
X_A,
y_A,
eval_set=[(X_B, y_B)],
eval_metric="rmse",
early_stopping_rounds=EARLY_STOPPING_ROUNDS,
verbose=0,
)
y_pred += model.predict(X_test.values)
y_pred /= N_REPEATS * N_SPLITS
The prediction is made of the average of the different out-of-fold predictions on the test set. We use the same cross-validation strategy as in the HPO process, in order to be consistent and achieve a similar level of error (0.8425081635928959).
In the following we are going to increment the submission file name and write the prediction as a CSV file :
sub_files = []
for root, dirs, files in os.walk("./"):
for file in files:
if file.startswith("submission_") and file.endswith(".csv"):
sub_files.append(file)
sub_files
if len(sub_files) == 0:
sub_files = ["submission_00.csv"]
sub_files.sort()
last_sub_file = sub_files[-1]
last_id = int(last_sub_file.split("_")[-1].split(".")[0])
curr_id = str(last_id + 1).zfill(2)
curr_sub_fn = "submission_" + curr_id + ".csv" # file name
test_df["target"] = y_pred
test_df[["target"]].to_csv(curr_sub_fn)
Now let’s submit :
!kaggle competitions submit -c tabular-playground-series-feb-2021 -f {curr_sub_fn} -m {curr_sub_fn}
100%|██████████████████████████████████████| 4.73M/4.73M [00:04<00:00, 1.04MB/s]
Successfully submitted to Tabular Playground Series - Feb 2021
Here is a capture of the leaderboard web page :
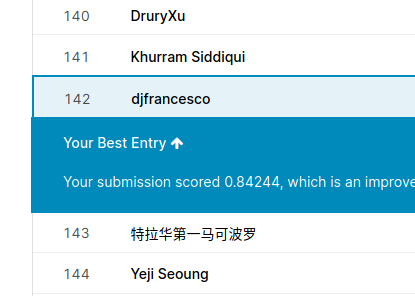
Not so bad, the public leaderboard score of the submission is 0.84244 (rank 142 / 826). Of course there would be a lot of work to do to improve this score (EDA, FE, other algorithms, stacking, magic tricks, …).