A Parallel loop in Python with Joblib.Parallel
The goal of this post is to perform an embarrassingly parallel loop in Python, with the same code running on different platforms (Linux and Windows). From wikipedia, here is a definition of embarassingly parallel:
In parallel computing, an embarrassingly parallel workload or problem […] is one where little or no effort is needed to separate the problem into a number of parallel tasks. This is often the case where there is little or no dependency or need for communication between those parallel tasks, or for results between them.
In the following, we are going to parallelize a loop with independent iterations. More specifically, we have list of natural numbers and want to check each number for primality.
Imports
from joblib import Parallel, delayed
from numba import jit
import numpy as np
import pandas as pd
import perfplot
import primesieve
Computations are performed on a laptop with an 8 cores Intel(R) i7-7700HQ CPU @ 2.80GHz running Linux. Package versions:
Python implementation: CPython
Python version : 3.9.10
IPython version : 8.0.1
perfplot : 0.10.1
pandas : 1.4.0
numpy : 1.21.5
primesieve: 2.3.0
The Parallel loop
Let’s say that we have a list of k natural numbers $\left[ n_1, n_2, …, n_k \right]$. We want to send roughly equal amounts of work to n_jobs processes and gather a list with all the k boolean results. For example, with k=16 and n_jobs=4:
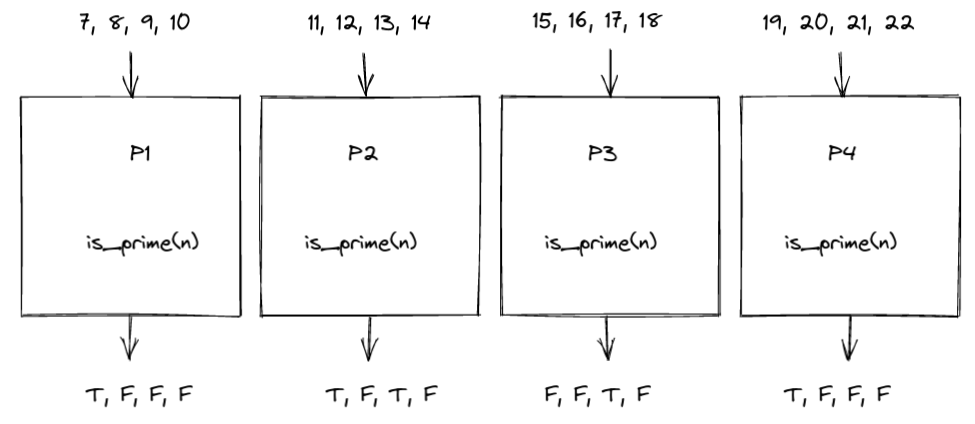
A Simple primality test
A primality test is an algorithm for determining whether an input number is prime. We are going to proceed with an old-school trial division method. For a given positive integer $n$, we check whether it can be divided by a smaller integer $p$. Also, we are going to perform two simple optimizations:
- If $n$ is not prime, it can be written as a product $n=p \, q$, and we obviously cannot have both $p$ and $q$ larger than $\sqrt{n}$. So we only test if $n$ can be divided by $p$ smaller or equal to $\sqrt{n}$. For example, when testing 25 for primality, we would check if 2, 3 or 5 are divisors. Numbers between 9 and 24 are either prime or multiple of 2 or 3. Numbers between 4 and 8 are either prime or even.
- Any integer $p \geq 5$ can be written as a multiple of 6 plus an integer $i$ between -1 and 4: $p=6k+i$, with $k \geq 1$ and $i \in \lbrace -1, 0, 1 , 2, 3, 4 \rbrace$. If $ i \in \lbrace 0, 3 \rbrace$ then $p$ is a multiple of 3, and if $i \in \lbrace 2, 4\rbrace$ then $p$ is a multiple of 2. So if we already made sure that $n$ is not a multiple of 2 or 3, we only need to check if $n$ can be divided by $p=6 \, k \pm 1$.
To summarize, we need to:
- deal first with $n \leq 3$
- check if $n>3$ is a multiple of 2 or 3.
- check if $p$ divides $n$ for $p = 6k \pm 1$ with $k \geq 1$ and $p \leq \sqrt{n}$. Note that we start here with $p=5$.
Here is a Python implementation of this test (here is the reference):
def is_prime(n: int) -> bool:
if n <= 3:
return n > 1
if (np.mod(n, 2) == 0) or (np.mod(n, 3) == 0):
return False
sqrt_n = int(np.floor(np.sqrt(n)))
p = 5
while p <= sqrt_n:
if (np.mod(n, p) == 0) or (np.mod(n, p + 2) == 0):
return False
p += 6
return True
Let’s check that it is correct with the list of prime numbers up to N=1000. We use the pyprimesieve package to computer a reference array of primes not larger than N:
N = 1_000
prime_indices_ref = np.array(primesieve.primes(N))
prime_indices_ref[-10:]
array([937, 941, 947, 953, 967, 971, 977, 983, 991, 997], dtype=uint64)
Then we also compute this list of primes using is_prime() and check it is equal to the above reference array:
is_prime_vec = list(map(is_prime, range(N + 1)))
prime_indices = np.where(is_prime_vec)[0]
np.testing.assert_array_equal(prime_indices, prime_indices_ref)
The is_prime function seems to work fine, but let’s try to accelerate it with Numba!
Numba
We just add the magic jit decorator to the function and perform the compilation by calling it once:
@jit(nopython=True, fastmath=True)
def is_prime_numba(n: int) -> bool:
if n <= 3:
return n > 1
if (np.mod(n, 2) == 0) or (np.mod(n, 3) == 0):
return False
sqrt_n = int(np.floor(np.sqrt(n)))
p = 5
while p <= sqrt_n:
if (np.mod(n, p) == 0) or (np.mod(n, p + 2) == 0):
return False
p += 6
return True
is_prime_numba(1234)
False
In order to measure the performance amelioration, we create a random number generator and generate an array of 1000 random integers. We then compare the is_prime and is_prime_numba functions by applying them to a range of integer arrays.
SD = 124
rng = np.random.default_rng(seed=SD)
SIZE = 1_000
out = perfplot.bench(
setup=lambda n: rng.integers(
np.power(10, n),
np.power(10, n + 1),
SIZE,
dtype=int,
endpoint=True,
),
kernels=[
lambda numbers: list(map(is_prime, numbers)),
lambda numbers: list(map(is_prime_numba, numbers)),
],
labels=["is_prime", "is_prime_numba"],
n_range=range(3, 13),
)
df = pd.DataFrame(data=out.timings_s, columns=out.n_range, index=out.labels).T
ax = df.plot(figsize=(12, 12), logy=True)
_ = ax.set(
title="Sequential acceleration with Numba",
xlabel="log10(n)",
ylabel=f"Runtime[s] (SIZE = {SIZE})",
)
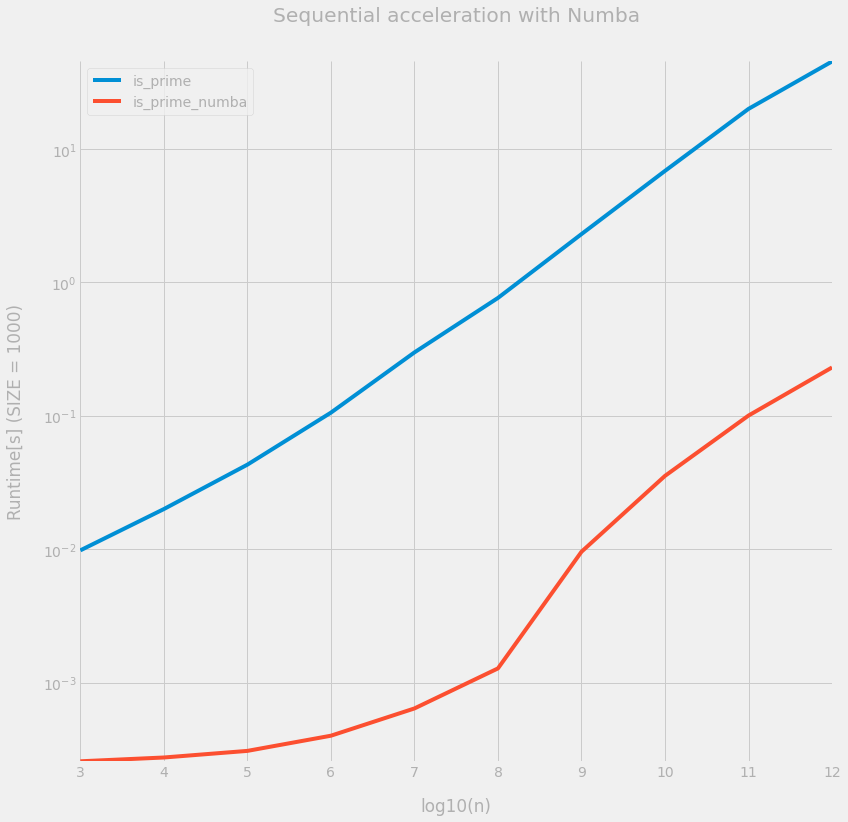
OK this is much faster with Numba. We are now ready to parallelize the loop on the integer array.
Parallel loop with Joblib
We are going to use joblib with the default loky backend. Loky is a cross-platform and cross-version implementation of the ProcessPoolExecutor class of concurrent.futures. One of its main features is (from Loky’s github repository):
No need for
if __name__ == "__main__":in scripts: thanks to the use of cloudpickle to call functions defined in the main module, it is not required to protect the code calling parallel functions under Windows.
This might be useful when writing a cross-platform library. We won’t go much into details about how new processes are created with multiprocessing in Linux and Windows systems, but here is for example a post describing this issue, written by Aquiles Carattino on the pythonforthelab website.
The current code is working with loky and multiprocessing on both Linux and Windows, but this is because it is running in a JupyterLab notebook. It would get trickier for a Python module on Windows with multiprocessing to avoid recursive spawning of subprocesses.
Also, note that is possible to use the dask backend on a distributed cluster.
Let’s start by generating an array of rather large integers:
LOW = int(1e12)
HIGH = int(1e13)
numbers = rng.integers(LOW, HIGH, SIZE, dtype=int, endpoint=True)
numbers[:5]
array([4229485319711, 6682422302023, 6589525053365, 7499770743267,
3694428446242])
We define a sequential function that applies is_prime_numba to all the array, returning a Pandas dataframe:
def is_prime_array_seq(numbers):
results = list(map(is_prime_numba, numbers))
res_df_seq = pd.DataFrame(
list(zip(numbers, results)), columns=["number", "is_prime"]
).set_index("number")
res_df_seq.sort_index(inplace=True)
return res_df_seq
res_df_seq = is_prime_array_seq(numbers)
res_df_seq.head(3)
| is_prime | |
|---|---|
| number | |
| 1010118466699 | True |
| 1010316874298 | False |
| 1011838077604 | False |
Now the parallel version is_prime_array_par:
def is_prime_array_par(numbers, n_jobs=8, batch_size=100, backend="loky"):
results = Parallel(
n_jobs=n_jobs, batch_size=batch_size, backend=backend, verbose=0
)(delayed(is_prime_numba)(n) for n in numbers)
res_df_par = pd.DataFrame(
list(zip(numbers, results)), columns=["number", "is_prime"]
).set_index("number")
res_df_par.sort_index(inplace=True)
return res_df_par
res_df_par = is_prime_array_par(numbers)
res_df_par.head(3)
| is_prime | |
|---|---|
| number | |
| 1010118466699 | True |
| 1010316874298 | False |
| 1011838077604 | False |
pd.testing.assert_frame_equal(res_df_seq, res_df_par)
Let’s compare the execution time of the sequential with the parallel version.
out = perfplot.bench(
setup=lambda n: rng.integers(
np.power(10, n),
np.power(10, n + 1),
SIZE,
dtype=int,
endpoint=True,
),
kernels=[
lambda numbers: is_prime_array_seq(numbers),
lambda numbers: is_prime_array_par(numbers, n_jobs=1),
lambda numbers: is_prime_array_par(numbers, n_jobs=2),
lambda numbers: is_prime_array_par(numbers, n_jobs=4),
lambda numbers: is_prime_array_par(numbers, n_jobs=8),
],
labels=["seq", "par_1", "par_2", "par_4", "par_8"],
n_range=range(12, 18),
)
df = pd.DataFrame(data=out.timings_s, columns=out.n_range, index=out.labels).T
ax = df.plot(figsize=(12, 12), logy=True)
_ = ax.set(
title="Parallel acceleration with Joblib",
xlabel="log10(n)",
ylabel=f"Runtime[s] (SIZE = {SIZE})",
)
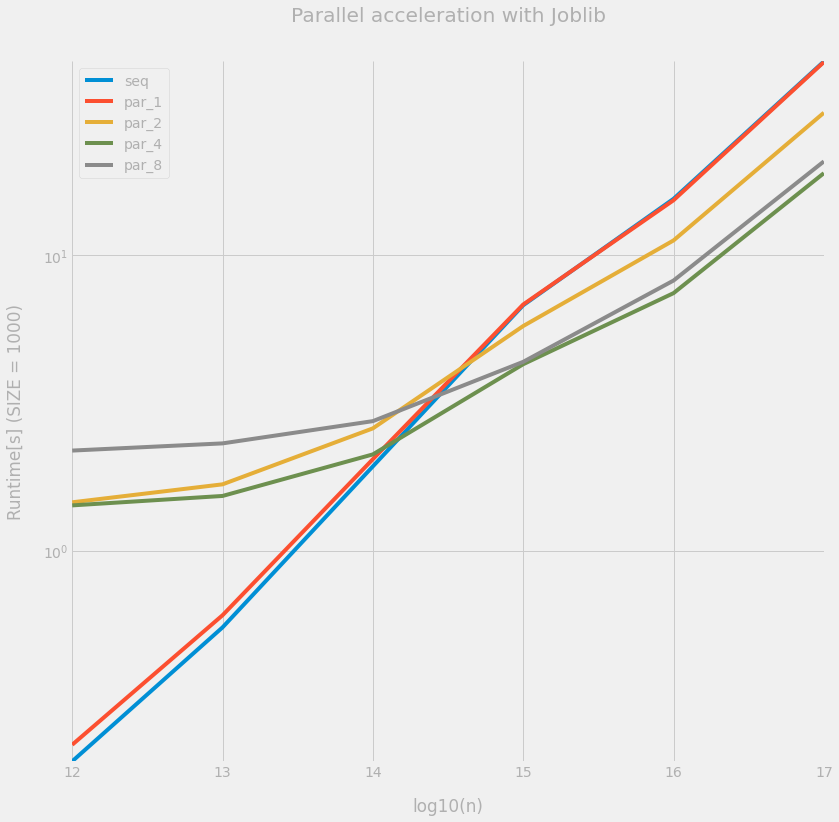
We can observe that the overhead of the parallelization is really significant: in the present case, it is only when $n \geq 10^{15}$ that the parallel version is faster than the sequential one, due to the heavier computational burden per worker. Also, n_jobs=4 is faster than n_jobs=8, probably because some other jobs were running on the laptop.
Conclusion
Thanks to Joblib with the loky backend, it is fairly easy to run an efficient embarrassingly parallel loop in Python. Furthermore, the same code is going to work on both Linux and Windows systems. However, one should make sure that the task distributed to each worker is large enough, so that the overhead induced by the pickling operation (happening when creating child processes) is negligible with regards to the worker computational load.