Loading data from PostgreSQL to Pandas with ConnectorX
ConnectorX is a library, written in Rust, that enables fast and memory-efficient data loading from various databases to different dataframes. We refer to this interesting paper, in which the authors provide a detailed analysis of the pandas.read_sql function:
Wang, Xiaoying, et al. ConnectorX: Accelerating Data Loading From Databases to Dataframes. 2021
This lead to some surprises:
A surprising finding is that the majority of the time is actually spent on the client side rather than on the query execution or the data transfer.
Most of the time is spent on deserialization and conversion to dataframe. So they tried to optimize these two steps, and also implemented an efficient parallelization based on query partitioning.
In the present post, we want to try the different ways, including with ConnectorX, to load some data with Python, from PostgreSQL to Pandas. Here are the different strategies tested:
- SQLAlchemy + psycopg2
- SQLAlchemy + psycopg2 by chunks
- pyodbc
- turbodbc + arrow
- ConnectorX + pandas
- ConnectorX + arrow
- ConnectorX + modin
- ConnectorX + dask
- ConnectorX + polars
- ConnectorX + pandas with 2 partitions
- ConnectorX + pandas with 4 partitions
- ConnectorX + pandas with 8 partitions
The Data
We have a table with fake data, created in Python with the Faker package, and stored as a Pandas dataframe df. Initially, this table has been loaded into PostgreSQL with the following piece of code, assumine the SQL engine has also already been created:
from sqlalchemy.types import (
Boolean,
Date,
DateTime,
Float,
Integer,
SmallInteger,
String,
Unicode,
)
l_name = 100
l_job = 200
l_email = 40
l_company = 150
l_industry = 150
l_city = 50
l_state = 50
l_zipcode = 15
dtypes = {
"name": Unicode(length=l_name),
"job": Unicode(length=l_job),
"birthdate": Date(),
"email": String(length=l_email),
"last_connect": DateTime(timezone=False),
"company": Unicode(length=l_company),
"industry": Unicode(length=l_industry),
"city": Unicode(length=l_city),
"state": Unicode(length=l_state),
"zipcode": String(length=l_zipcode),
"netNew": Boolean(),
"sales1_rounded": Integer(),
"sales2_decimal": Float(),
"sales2_rounded": Integer(),
"priority": SmallInteger(),
}
df.to_sql(
"faker_s1000000i",
engine,
if_exists="replace",
index=True,
index_label="index",
dtype=dtypes,
)
Here is a SELECT on the table from pgAdmin:
The table has a million rows and 16 columns. The query used to load the data is basic:
QUERY = 'SELECT * FROM "faker_s1000000i"'
We are going to compare the loading time using different drivers and methods. We do not measure the peak memory, but just the elapsed time for loading the data into Pandas. Database server and client are on the same machine (my laptop).
Imports
import urllib
import connectorx as cx
import pandas as pd
import psycopg2
import pyodbc
import ray
import turbodbc
from sqlalchemy import create_engine
ray.init(ignore_reinit_error=True)
Package versions:
Python : 3.9.12
psycopg2 : 2.9.3
sqlalchemy: 1.4.35
numpy : 1.22.3
ray : 1.11.0
connectorx: 0.2.5
turbodbc : 4.5.3
pyodbc : 4.0.32
pandas : 1.4.1
Credentials
pg_username = "****"
pg_password = "****"
pg_server = "localhost"
pg_port = 5432
pg_database = "pgdb"
SQLAlchemy + psycopg2 (sqlalchemy_psychopg2)
This is the most common way for loading data from Postgres into Pandas.
connect_string = f"postgresql+psycopg2://{pg_username}:{urllib.parse.quote_plus(pg_password)}@{pg_server}:{pg_port}/{pg_database}"
engine_psycopg2 = create_engine(connect_string)
%%time
with engine_psycopg2.connect() as connection:
df = pd.read_sql(sql=QUERY, con=connection)
CPU times: user 7.06 s, sys: 755 ms, total: 7.81 s
Wall time: 8.54 s
assert isinstance(df, pd.core.frame.DataFrame)
assert df.shape == (1000000, 16)
SQLAlchemy + psycopg2 by chunks (sqlalchemy_psychopg2_chunks)
Now let’s imagine that we want to reduce the memory usage of the previous process by loading the data by chunks. This code is inspired by the ConnectorX github repository, where a lot of usefull benchmark code can be found.
%%time
chunksize = 1_000
engine = create_engine(connect_string)
connection = engine.connect().execution_options(
stream_results=True, max_row_buffer=chunksize
)
dfs = []
for df in pd.read_sql(QUERY, connection, chunksize=chunksize):
dfs.append(df)
df = pd.concat(dfs)
connection.close()
CPU times: user 9.39 s, sys: 428 ms, total: 9.82 s
Wall time: 11 s
assert isinstance(df, pd.core.frame.DataFrame)
assert df.shape == (1000000, 16)
pyodbc (pyodbc)
The official PostgreSQL ODBC driver is used here with default settings.
connection = pyodbc.connect(dsn="PostgreSQL")
%%time
df = pd.read_sql(sql=QUERY, con=connection)
UserWarning: pandas only support SQLAlchemy connectable(engine/connection) ordatabase string URI or sqlite3 DBAPI2 connectionother DBAPI2 objects are not tested, please consider using SQLAlchemy
warnings.warn(
CPU times: user 16.9 s, sys: 723 ms, total: 17.6 s
Wall time: 17.7 s
connection.close()
assert isinstance(df, pd.core.frame.DataFrame)
assert df.shape == (1000000, 16)
Turbodbc + arrow (turbodbc_arrow)
Here we do not use Pandas’read_sql function but Apache Arrow along with Turbodbc. See this blog post for more details: https://arrow.apache.org/blog/2017/06/16/turbodbc-arrow/
connection = turbodbc.connect(dsn="PostgreSQL")
%%time
cursor = connection.cursor()
cursor.execute(QUERY)
data = cursor.fetchallarrow()
df = data.to_pandas(split_blocks=False, date_as_object=True)
CPU times: user 7.17 s, sys: 374 ms, total: 7.55 s
Wall time: 7.8 s
The type of the data returned above by fetchallarrow() is an Arrow table, this is why it is later converted to a Pandas dataframe.
connection.close()
assert isinstance(df, pd.core.frame.DataFrame)
assert df.shape == (1000000, 16)
ConnectorX
We are going to connect the data source with the following connection string URI and use the cx.read_sql function:
connect_string = f"postgres://{pg_username}:{urllib.parse.quote_plus(pg_password)}@{pg_server}:{pg_port}/{pg_database}"
Pandas (cx_pandas)
%%time
df = cx.read_sql(conn=connect_string, query=QUERY, return_type="pandas")
CPU times: user 3.71 s, sys: 278 ms, total: 3.99 s
Wall time: 3.96 s
assert isinstance(df, pd.core.frame.DataFrame)
assert df.shape == (1000000, 16)
Arrow (cx_arrow)
%%time
table = cx.read_sql(conn=connect_string, query=QUERY, return_type="arrow")
CPU times: user 3.2 s, sys: 237 ms, total: 3.43 s
Wall time: 3.35 s
type(table)
pyarrow.lib.Table
Since the returning type is not a Pandas dataframe but an Arrow table, we need to convert it (and take the conversion time into account):
%%time
df = table.to_pandas()
CPU times: user 1.21 s, sys: 209 ms, total: 1.42 s
Wall time: 1.4 s
assert isinstance(df, pd.core.frame.DataFrame)
assert df.shape == (1000000, 16)
Modin (cx_modin)
%%time
df = cx.read_sql(conn=connect_string, query=QUERY, return_type="modin")
UserWarning: The pandas version installed 1.4.2 does not match the supported pandas version in Modin 1.4.1. This may cause undesired side effects!
UserWarning: Distributing <class 'pandas.core.frame.DataFrame'> object. This may take some time.
CPU times: user 6.09 s, sys: 629 ms, total: 6.72 s
Wall time: 6.78 s
type(df)
modin.pandas.dataframe.DataFrame
Here we need to convert from a Modin to a Pandas dataframe:
%%time
df = df._to_pandas()
CPU times: user 1.28 s, sys: 266 ms, total: 1.55 s
Wall time: 1.53 s
assert isinstance(df, pd.core.frame.DataFrame)
assert df.shape == (1000000, 16)
Dask (cx_dask)
%%time
df = cx.read_sql(conn=connect_string, query=QUERY, return_type="dask")
CPU times: user 5.4 s, sys: 528 ms, total: 5.92 s
Wall time: 5.88 s
type(df)
dask.dataframe.core.DataFrame
Each partition in a Dask DataFrame is a Pandas DataFrame. Since we are presumably using a single partition in the present case, converting to Pandas is almost instentaneous:
%%time
df = df.compute()
CPU times: user 1.53 ms, sys: 86 µs, total: 1.62 ms
Wall time: 1.15 ms
assert isinstance(df, pd.core.frame.DataFrame)
assert df.shape == (1000000, 16)
Polars (cx_polars)
%%time
df = cx.read_sql(conn=connect_string, query=QUERY, return_type="polars")
CPU times: user 3.34 s, sys: 255 ms, total: 3.59 s
Wall time: 3.51 s
type(df)
polars.internals.frame.DataFrame
Again, we need to convert from a Polars to a Pandas dataframe :
%%time
df = df.to_pandas()
CPU times: user 848 ms, sys: 149 ms, total: 997 ms
Wall time: 982 ms
assert isinstance(df, pd.core.frame.DataFrame)
assert df.shape == (1000000, 16)
Pandas 2 partitions (cx_pandas_2)
%%time
df = cx.read_sql(conn=connect_string, query=QUERY, return_type="pandas", partition_on="index", partition_num=2)
CPU times: user 4.77 s, sys: 596 ms, total: 5.37 s
Wall time: 2.88 s
assert isinstance(df, pd.core.frame.DataFrame)
assert df.shape == (1000000, 16)
Pandas 4 partitions (cx_pandas_4)
%%time
df = cx.read_sql(conn=connect_string, query=QUERY, return_type="pandas", partition_on="index", partition_num=4)
CPU times: user 5.95 s, sys: 657 ms, total: 6.61 s
Wall time: 2.2 s
assert isinstance(df, pd.core.frame.DataFrame)
assert df.shape == (1000000, 16)
Pandas 8 partitions (cx_pandas_8)
%%time
df = cx.read_sql(conn=connect_string, query=QUERY, return_type="pandas", partition_on="index", partition_num=8)
CPU times: user 5.72 s, sys: 689 ms, total: 6.41 s
Wall time: 2.28 s
assert isinstance(df, pd.core.frame.DataFrame)
assert df.shape == (1000000, 16)
Comparison
We did not include here the code associated with time measurements. Basically, for each strategy, the loading process is repeated 5 times and only the best elapsed time is kept. Here is the resulting table:
| read_sql | to_frame | total | |
|---|---|---|---|
| pyodbc | 17.79901 | 0.00000 | 17.79901 |
| sqlalchemy_psycopg2_chunks | 10.87270 | 0.00000 | 10.87270 |
| sqlalchemy_psycopg2 | 9.14140 | 0.00000 | 9.14140 |
| cx_modin | 6.42423 | 1.54774 | 7.97197 |
| turbodbc_arrow | 6.36288 | 0.96034 | 7.32322 |
| cx_dask | 5.29873 | 0.00043 | 5.29916 |
| cx_arrow | 3.16782 | 1.11604 | 4.28385 |
| cx_polars | 3.23051 | 0.97591 | 4.20642 |
| cx_pandas | 3.95265 | 0.00000 | 3.95265 |
| cx_pandas_2 | 2.48966 | 0.00000 | 2.48966 |
| cx_pandas_8 | 2.12081 | 0.00000 | 2.12081 |
| cx_pandas_4 | 1.93755 | 0.00000 | 1.93755 |
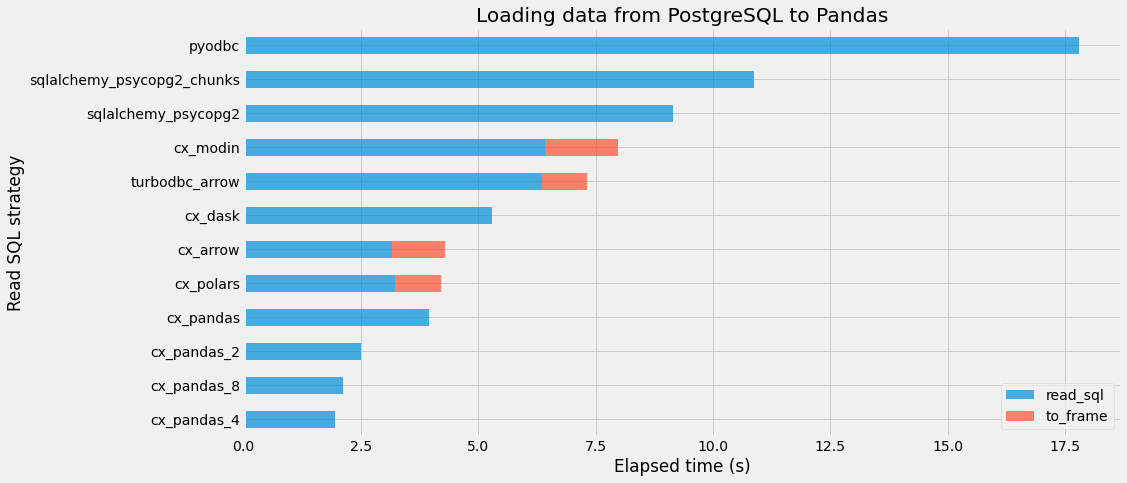
read_sql corrrespond to the elapsed time spent in loading the data into a container, while to_frame is the elapsed time spent in converting this container into a Pandas dataframe, if necessary.
We can see that ConnectorX is an efficient tool. We did not measure memory efficiency in this post, but it is supposed to be interesting as well. The supported data sources are the following (from the documentation): Postgres, Mysql, Mariadb (through mysql protocol), Sqlite, Redshift (through postgres protocol), Clickhouse (through mysql protocol), SQL Server, Azure SQL Database (through mssql protocol), Oracle. It would be great if more data sources could be supported, such as SAP Hana for example!
Although we loaded the data into a Pandas dataframe in the present post, we could also imagine to use ConnectorX along with Polars dataframes to perform some data analysis tasks.