Streaming data from PostgreSQL to a CSV file
Update Jan 25, 2024 - Added FastBCP.
In this post, we explore the process of streaming data from a PostgreSQL database to a CSV file using Python. The primary goal is to avoid loading the entire dataset into memory, enabling a more scalable and resource-efficient approach.
We’ll try out various Python libraries and techniques and evaluate their performance in terms of elapsed time and memory:
- Pandas
- Pandas + PyArrow
- Turbodbc + PyArrow
- Psycopg2
- ADBC + PyArrow
- DuckDB
Also, we will include a fast in-house extraction tool written in C#:
- FastBCP
System and package versions
We are operating on Python version 3.11.7 and running on a Linux x86_64 machine.
adbc-driver-postgresql : 0.8.0
duckdb : 0.9.2
pandas : 2.1.4
psycopg2 : 2.9.9
pyarrow : 14.0.2
sqlalchemy : 2.0.24
turbodbc : 4.9.0
Imports
import csv
import json
import os
import time
import adbc_driver_postgresql.dbapi
import duckdb
import pandas as pd
import psycopg2
import pyarrow as pa
import pyarrow.csv
from sqlalchemy import create_engine
from turbodbc import connect, make_options
# file paths for the CSV output and database credentials,
csv_file_path = os.path.abspath("./test_lineitem.csv")
credentials_file_path = os.path.abspath("./credentials.json")
While the credentials retrieved below serve the classic connection method, we also employ an ODBC connection later on, with a Data Source Name (DSN).
with open(credentials_file_path, "r") as json_file:
creds = json.load(json_file)
print("creds keys: ", list(creds.keys()))
creds keys: ['username', 'password', 'server', 'port', 'database']
# initialization of the dict storing the elapsed time for different approaches
elapsed_time = {}
Query
The TPC-H benchmark stands as a widely-utilized metric for assessing the performance of database systems. The data can be created using pre-determined database sizes, referred to as scale factors. In this context, we are utilizing a scale factor of 10, focusing on the largest table among the 8 TPCH tables: lineitem. This table comprises approximately 60 million (59,986,052) rows and 16 columns. Below is a glimpse of the first 10 rows of the table:
┌────────────┬───────────┬───────────┬───┬────────────┬──────────────────────┐
│ l_orderkey │ l_partkey │ l_suppkey │ … │ l_shipmode │ l_comment │
│ int64 │ int64 │ int64 │ │ varchar │ varchar │
├────────────┼───────────┼───────────┼───┼────────────┼──────────────────────┤
│ 1 │ 1551894 │ 76910 │ … │ TRUCK │ to beans x-ray car… │
│ 1 │ 673091 │ 73092 │ … │ MAIL │ according to the … │
│ 1 │ 636998 │ 36999 │ … │ REG AIR │ ourts cajole above… │
│ 1 │ 21315 │ 46316 │ … │ AIR │ s cajole busily ab… │
│ 1 │ 240267 │ 15274 │ … │ FOB │ the regular, regu… │
│ 1 │ 156345 │ 6348 │ … │ MAIL │ rouches. special │
│ 2 │ 1061698 │ 11719 │ … │ RAIL │ re. enticingly reg… │
│ 3 │ 42970 │ 17971 │ … │ AIR │ s cajole above the… │
│ 3 │ 190355 │ 65359 │ … │ RAIL │ ecial pinto beans.… │
│ 3 │ 1284483 │ 34508 │ … │ SHIP │ e carefully fina │
├────────────┴───────────┴───────────┴───┴────────────┴──────────────────────┤
│ 10 rows 16 columns (5 shown) │
└────────────────────────────────────────────────────────────────────────────┘
Additionally, we instruct the SQL engine to sort the table by the l_orderkey column:
sql = "SELECT * FROM tpch_10.lineitem ORDER BY l_orderkey"
All the following approaches will employ the same SQL query. Let’s start with the first technique.
Pandas
The code creates a SQLAlchemy engine for PostgreSQL using the psycopg2 driver, specifying the connection details from the credentials.
The stream_results=True and max_row_buffer=chunk_size connection options are used to stream the results and limit the number of rows buffered in memory.
The code then iterates through the chunks obtained from the SQL query using pd.read_sql from Pandas. It writes each chunk to a CSV file in append mode after the first chunk.
%%time
start_time_step = time.perf_counter()
engine = create_engine(
f"postgresql+psycopg2://{creds['username']}:{creds['password']}@{creds['server']}:{creds['port']}/{creds['database']}"
)
chunk_size = 100_000
with engine.connect().execution_options(
stream_results=True, max_row_buffer=chunk_size
) as conn:
first_chunk = True
mode = "w"
for df in pd.read_sql(sql=sql, con=conn, chunksize=chunk_size):
df.to_csv(
csv_file_path, mode=mode, sep=';', header=first_chunk, quoting=csv.QUOTE_ALL, index=False
)
if first_chunk:
mode = "a"
first_chunk = False
elapsed_time["Pandas"] = time.perf_counter() - start_time_step
CPU times: user 6min 26s, sys: 8.84 s, total: 6min 35s
Wall time: 7min 17s
The CSV file containing the data from the PostgreSQL table, occupies approximately 10 gigabytes of disk space. Note that if we don’t activate the stream_results option, the data is fully loaded into memory by the read_sql statement, even with smaller chunks. This does not fit in the available RAM on the computer.
Next we display the first three and last three rows of the generated CSV file:
!head -n 3 {csv_file_path}
"l_orderkey";"l_partkey";"l_suppkey";"l_linenumber";"l_quantity";"l_extendedprice";"l_discount";"l_tax";"l_returnflag";"l_linestatus";"l_shipdate";"l_commitdate";"l_receiptdate";"l_shipinstruct";"l_shipmode";"l_comment"
"1";"1551894";"76910";"1";"17.0";"33078.94";"0.04";"0.02";"N";"O";"1996-03-13";"1996-02-12";"1996-03-22";"DELIVER IN PERSON ";"TRUCK ";"to beans x-ray carefull"
"1";"673091";"73092";"2";"36.0";"38306.16";"0.09";"0.06";"N";"O";"1996-04-12";"1996-02-28";"1996-04-20";"TAKE BACK RETURN ";"MAIL ";" according to the final foxes. qui"
!tail -n 3 {csv_file_path}
"60000000";"118838";"93842";"5";"28.0";"51991.24";"0.0";"0.08";"N";"O";"1997-09-29";"1997-11-06";"1997-09-30";"COLLECT COD ";"SHIP ";"regular foxes among the even depths use "
"60000000";"1294851";"19864";"6";"48.0";"88597.92";"0.03";"0.07";"N";"O";"1997-11-28";"1997-10-05";"1997-12-06";"COLLECT COD ";"MAIL ";"ual asymptotes wake af"
"60000000";"558286";"33302";"7";"12.0";"16131.12";"0.02";"0.05";"N";"O";"1997-10-09";"1997-10-27";"1997-10-21";"COLLECT COD ";"REG AIR ";"ickly according to the furiousl"
Also we check that the l_orderkey column is sorted:
check_df = pd.read_csv(csv_file_path, delimiter=";", usecols=["l_orderkey"])
assert check_df.l_orderkey.is_monotonic_increasing
Pandas + PyArrow
In this section we use the Pandas library with the pyarrow backend in combination with PyArrow for seamlessly handle Arrow data, i.e. write the CSV file. There should be no data copy between Pandas and PyArrow.
We are still using a SQLAlchemy engine with the psycopg2 driver and the stream_results=True and max_row_buffer=chunk_size connection mode.
%%time
start_time_step = time.perf_counter()
engine = create_engine(
f"postgresql+psycopg2://{creds['username']}:{creds['password']}@{creds['server']}:{creds['port']}/{creds['database']}"
)
chunk_size = 100_000
write_options = pa.csv.WriteOptions(
include_header=True,
batch_size=2_048,
delimiter=";",
quoting_style="all_valid",
)
is_writer = False
with engine.connect().execution_options(
stream_results=True, max_row_buffer=chunk_size
) as conn:
for df in pd.read_sql(
sql=sql, con=conn, chunksize=chunk_size, dtype_backend="pyarrow"
):
table = pa.Table.from_pandas(df)
if not is_writer:
schema = table.schema
writer = pa.csv.CSVWriter(
sink=csv_file_path, schema=schema, write_options=write_options
)
is_writer = True
writer.write_table(table)
writer.close()
elapsed_time["Pandas+PyArrow"] = time.perf_counter() - start_time_step
CPU times: user 5min 8s, sys: 8.2 s, total: 5min 16s
Wall time: 5min 50s
check_df = pd.read_csv(csv_file_path, delimiter=";", usecols=["l_orderkey"])
assert check_df.l_orderkey.is_monotonic_increasing
Turbodbc + PyArrow
turbodbc is a module to access relational databases via the Open Database Connectivity (ODBC) interface. Unlike previous approaches, we establish the connection using the data source name (DSN). Notably, asynchronous I/O is activated during data retrieval, allowing the fetching of new result sets from the database in the background while Python processes the existing ones.
%%time
start_time_step = time.perf_counter()
options = make_options(
use_async_io=True,
prefer_unicode=True,
)
write_options = pa.csv.WriteOptions(
include_header=True,
batch_size=2048,
delimiter=";",
quoting_style="all_valid",
)
with connect(connection_string="dsn=PostgreSQL", turbodbc_options=options) as conn:
with conn.cursor() as cur:
_ = cur.execute(sql=sql)
is_writer = False
for table in cur.fetcharrowbatches():
if not is_writer:
schema = table.schema
writer = pa.csv.CSVWriter(
sink=csv_file_path, schema=schema, write_options=write_options
)
is_writer = True
writer.write_table(table)
writer.close()
elapsed_time["Turbodbc+PyArrow"] = time.perf_counter() - start_time_step
CPU times: user 2min 34s, sys: 6.08 s, total: 2min 40s
Wall time: 2min 4s
check_df = pd.read_csv(csv_file_path, delimiter=";", usecols=["l_orderkey"])
assert check_df.l_orderkey.is_monotonic_increasing
Psycopg2
Psycopg is a wrapper for the libpq C library, the official PostgreSQL client library. We are going to use COPY () TO STDOUT which streams data to the client, along with the copy_expert method.
%%time
start_time_step = time.perf_counter()
output_query = f"COPY ({sql}) TO STDOUT WITH (FORMAT CSV, HEADER true, DELIMITER ';', FORCE_QUOTE *)"
with psycopg2.connect(
host=creds["server"],
user=creds["username"],
password=creds["password"],
dbname=creds["database"],
port=creds["port"],
) as conn:
with conn.cursor() as cur:
with open(csv_file_path, "w") as f:
cur.copy_expert(output_query, f)
elapsed_time["Psycopg2"] = time.perf_counter() - start_time_step
CPU times: user 9.72 s, sys: 7.31 s, total: 17 s
Wall time: 51.9 s
check_df = pd.read_csv(csv_file_path, delimiter=";", usecols=["l_orderkey"])
assert check_df.l_orderkey.is_monotonic_increasing
ADBC + PyArrow
In this section, we leverage ADBC in conjunction with PyArrow to fetch data as Arrow batches and subsequently write it to a CSV file. ABDC stands for the Arrow Database Connectivity:
ADBC aims to provide a minimal database client API standard, based on Arrow, for C, Go, and Java (with bindings for other languages). Applications code to this API standard (in much the same way as they would with JDBC or ODBC), but fetch result sets in Arrow format (e.g. via the C Data Interface). They then link to an implementation of the standard: either directly to a vendor-supplied driver for a particular database, or to a driver manager that abstracts across multiple drivers. Drivers implement the standard using a database-specific API, such as Flight SQL.
The ADBC implementation utilized in this context is specifically designed for PostgreSQL. However, alternative implementations may provide support for databases such as DuckDB, Snowflake, or SQLite.
Data is fetched as Arrow batches and written with PyArrow.
%%time
start_time_step = time.perf_counter()
write_options = pa.csv.WriteOptions(
include_header=True,
delimiter=";",
quoting_style="all_valid",
)
uri = f"postgresql://{creds['username']}:{creds['password']}@{creds['server']}:{creds['port']}/{creds['database']}"
with adbc_driver_postgresql.dbapi.connect(uri) as conn:
with conn.cursor() as cur:
cur.execute(sql)
is_writer = False
for batch in cur.fetch_record_batch():
if not is_writer:
schema = batch.schema
writer = pa.csv.CSVWriter(
sink=csv_file_path, schema=schema, write_options=write_options
)
is_writer = True
writer.write_batch(batch)
writer.close()
elapsed_time["ADBC+PyArrow"] = time.perf_counter() - start_time_step
CPU times: user 1min 5s, sys: 5.95 s, total: 1min 11s
Wall time: 1min 11s
check_df = pd.read_csv(csv_file_path, delimiter=";", usecols=["l_orderkey"])
assert check_df.l_orderkey.is_monotonic_increasing
DuckDB
In this segment, we explore the integration of the in-process database DuckDB with its postgres extension, allowing for bidirectional data transfer between DuckDB and PostgreSQL:
The postgres extension allows DuckDB to directly read and write data from a running Postgres database instance. The data can be queried directly from the underlying Postgres database. Data can be loaded from Postgres tables into DuckDB tables, or vice versa.
To enable the PostgreSQL extension, a straightforward SQL command is employed: INSTALL postgres;. Following this installation, and the extension loading, the ATTACH command is utilized to make the PostgreSQL database accessible to DuckDB. In this process, an alias, db in this present case, is assigned to the database, and accordingly, SQL queries need to be adjusted to consider this alias. The SELECT query is modified as follows:
sql_duckdb = sql.replace("tpch_", "db.tpch_")
sql_duckdb
Now here call this query with the postgres_query function:
%%time
start_time_step = time.perf_counter()
conn = duckdb.connect()
conn.sql("INSTALL postgres;")
conn.sql("LOAD postgres;")
conn.sql(
f"""ATTACH 'dbname={creds["database"]} user={creds["username"]} password={creds["password"]} host={creds["server"]} port={creds["port"]}'
AS db (TYPE POSTGRES);"""
)
conn.sql(
f"""COPY (SELECT * FROM postgres_query('db', '{sql}'))
TO '{csv_file_path}' (HEADER, DELIMITER ';', force_quote *);"""
)
conn.close()
elapsed_time["DuckDB"] = time.perf_counter() - start_time_step
CPU times: user 1min 2s, sys: 5.17 s, total: 1min 7s
Wall time: 1min 7s
check_df = pd.read_csv(csv_file_path, delimiter=";", usecols=["l_orderkey"])
assert check_df.l_orderkey.is_monotonic_increasing
FastBCP
While our focus here is on Python tools, we added FastBCP as a reference regarding CPU and memory usage. FastBCP has been developed at Architecture & Performance. It is a command line tool, written in C#, that is compatible with any operating system where dotnet is installed. We used dotnet on Linux in the present case.
FastBCP employs parallel threads, reading data through multiple connections by partitioning SQL on the ‘l_orderkey’ column, using the “random” method. This approach results in distinct CSV files, later merged into a final output. It’s worth mentioning that due to its parallel settings, the resulting data in the CSV file may not be sorted. This is why the ORDER BY clause is removed from the query in this particular case. Also, the returned elapsed time take the merging phase into account.
For reference, here’s the Python script used:
import subprocess
start_time_step = time.perf_counter()
output_dir_path = os.path.basename(csv_file_path)
output_file_name = os.path.dirname(csv_file_path)
n_jobs = 8
delimiter = ";"
sql_fastbcp = "SELECT * FROM tpch_10.lineitem"
subprocess.run(
f"""./FastBCP -C pgcopy -S {creds['server']} -U "{creds['username']}" -X "{creds['password']}" -I tpch -D {output_dir_path} -o {output_file_name} -p {n_jobs} -q "{sql_fastbcp}" -m Random -n "." -d "{delimiter}" -t false -f "yyyy-MM-dd HH:mm:ss" -e "UTF-8" -c "(L_ORDERKEY/10000000)::int" -M true""",
shell=True,
check=True,
)
elapsed_time["FastBCP"] = time.perf_counter() - start_time_step
Notice that 8 jobs were configured for this execution.
Results
et_df = pd.DataFrame.from_dict(elapsed_time, orient="index", columns=["Elapsed time (s)"])
ax = et_df.sort_values(by="Elapsed time (s)").plot.barh(
alpha=0.7, legend=False, figsize=(6, 4)
)
_ = ax.set(title="TPCH-SF10 lineitem table CSV extract", xlabel="Elapsed time (s)")
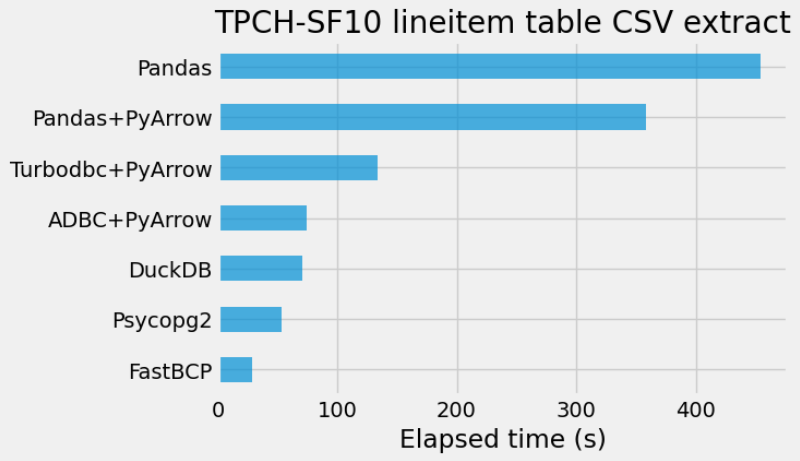
Let’s look at the memory usage of each method with the memory_profiler package. This is done outside of Jupyter using Python scripts.
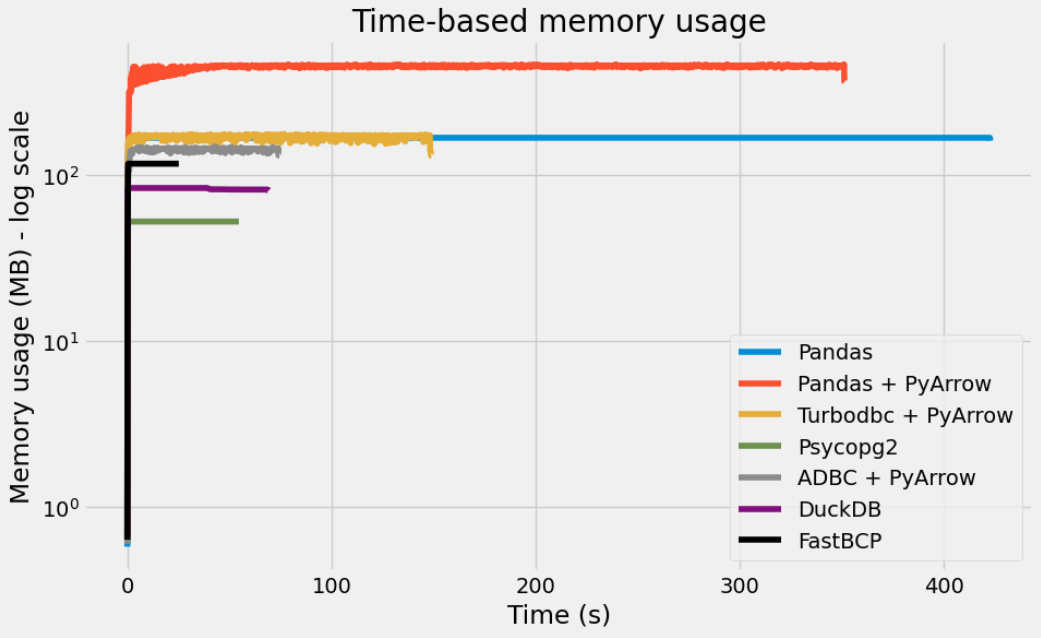
So, overall, Psycopg2, DuckDB and ADBC + PyArrow all achieve a high level of performance.
While we briefly included FastBCP for a reference comparison, we did not delve into extensive details. However, in the near future, we plan to focus on it in a dedicated post, thoroughly examining its behavior and performance in the context of data extraction.
Additionally, it’s worth mentioning that we did not manage to employ Polars for the streaming extraction, leading to an out-of-memory error.
Also, it appears that ConnectorX currently lacks support for retrieving results as Arrow batches or any type of chunks, making it unfit for this task.