Forward and reverse stars in Cython
This notebook is the following of a previous one, where we looked at the forward and reverse star representations of a sparse directed graph in pure Python:
Forward and reverse star representation of a digraph.
The motivation is to access the outgoing or incoming edges of graph nodes, as well as the associated edge attributes, in an efficient manner. The type of networks that we are interested in are road networks, which usually have a rather low mean degree. The use case that we have in mind are shortest path algorithms. In the present case, we only deal with a single edge attribute: a weight with float type. However we could use more attributes, such as in some shortest hyperpath algorithms:
- link travel time
- maximum link delay
- probability that the link is used in the hyperpath
In the following, we implement some code in Cython. What is Cython? Here is the description from the Cython web site:
Cython is an optimising static compiler for both the Python programming language and the extended Cython programming language (based on Pyrex). It makes writing C extensions for Python as easy as Python itself.
We refer to the Cython documentation for an in-depth presentation of the language.
In this note book, we are going to implement three different graph representations in Cython:
- Forward and reverse stars with NumPy 1D arrays
- Forward star with a NumPy structured array
- Adjacency list with pointers and C structs

Imports
import os
import numpy as np
import pandas as pd
from scipy.sparse import coo_array
%load_ext cython
RS = 124 # random seed
Package versions:
Python version : 3.10.6
cython : 0.29.32
jupyterlab : 3.5.0
numpy : 1.23.4
pandas : 1.5.1
scipy : 1.9.3
We start by creating a very small network for testing purpose.
Create a small network
This is a network already used in the previous post. Note that this graph has some parallel edges, a loop and an isolated vertex.
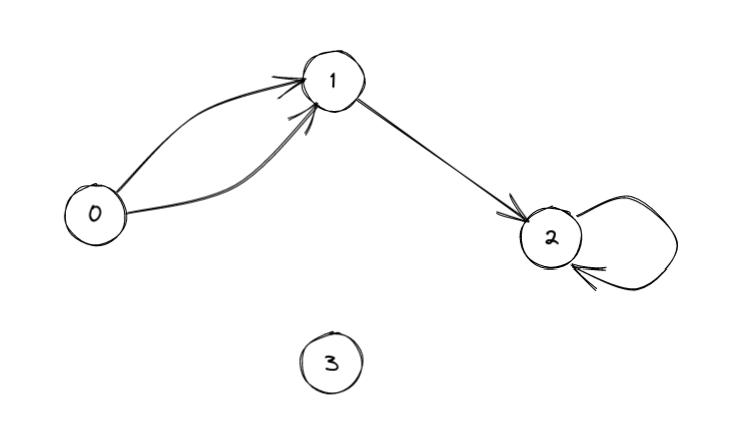
We store the network as an edge dataframe called edges_df_S:
tail_verts_S = np.array([0, 0, 1, 3], dtype=np.uint32)
head_verts_S = np.array([1, 1, 3, 3], dtype=np.uint32)
edge_weights_S = np.array([2, 1, 2, 3], dtype=np.float64)
edges_df_S = pd.DataFrame(
data={
"source": tail_verts_S,
"target": head_verts_S,
"weight": edge_weights_S,
}
)
vertex_count_S = int(edges_df_S[["source", "target"]].max().max() + 1)
edge_count_S = int(len(edges_df_S))
print(f"{edge_count_S} edges and {vertex_count_S} vertices")
edges_df_S[["source", "target"]] = edges_df_S[["source", "target"]].astype(np.uint32)
edges_df_S.head(4)
4 edges and 4 vertices
| source | target | weight | |
|---|---|---|---|
| 0 | 0 | 1 | 2.0 |
| 1 | 0 | 1 | 1.0 |
| 2 | 1 | 3 | 2.0 |
| 3 | 3 | 3 | 3.0 |
This dataframe describing the graph edges has 3 columns. Each row represents an edge with associated source node, target node and edge weight.
edges_df_S.dtypes
source uint32
target uint32
weight float64
dtype: object
Load a larger network
We are going to load the USA road network, with 57708624 edges, from the 9th DIMACS implementation challenge. We saved it into an Apache Parquet file in a previous post. We load this Parquet file into a Pandas dataframe named edges_df_L.
An important feature of this network is that duplicated edges have been removed. This is why we can compare the CSR or CSC sparse formats implemented later in Cython with the one produced by SciPy.sparse.coo_array methods: .tocsr and .tocsc. Since the SciPy module has been designed for matrices and not graphs, it handles duplicate entries by summing them. Parallel edges correspond to multiple matrix entries at the same row-column location when considering the node-node adjacency matrix. So duplicated edges would be “merged into a single one” by these methods, with a larger edge weight, which is not what we want here.
%%time
DATA_DIR = "/home/francois/Data/Disk_1/"
reg = "USA"
network_file_path = os.path.join(
DATA_DIR, f"DIMACS_road_networks/{reg}/USA-road-t.{reg}.gr.parquet"
)
edges_df_L = pd.read_parquet(network_file_path)
edges_df_L.rename(
columns={"id_from": "source", "id_to": "target", "tt": "weight"}, inplace=True
)
edges_df_L[["source", "target"]] = edges_df_L[["source", "target"]].astype(np.uint32)
vertex_count_L = int(edges_df_L[["source", "target"]].max().max() + 1)
edge_count_L = int(len(edges_df_L))
print(f"{edge_count_L} edges and {vertex_count_L} vertices")
57708624 edges and 23947347 vertices
CPU times: user 6.15 s, sys: 2.84 s, total: 8.99 s
Wall time: 4.16 s
edges_df_L.dtypes
source uint32
target uint32
weight float64
dtype: object
Also, later we are going to walk randomly though each node of this USA network and use the forward/reverse stars to access all outgoing or incoming edges. So let’s create a random vector with a random perlmutation of all the node indices:
%%time
rng = np.random.default_rng(RS)
node_randseq_L = rng.permutation(vertex_count_L).astype(np.uint32)
print(node_randseq_L)
[16377060 22703014 16671266 ... 10845119 17193243 18514748]
CPU times: user 790 ms, sys: 23.8 ms, total: 814 ms
Wall time: 811 ms
Convert to SciPy COO format for validation purpose
Next we convert this large graph into scipy.sparse Coordinate list (COO) format. We will later use it to check the result of our Cython Forward and Reverse star functions. Again, we do not convert the small graph because of the parallel edges.
%%time
sp_coo_L = coo_array(
(
edges_df_L["weight"].values,
(edges_df_L["source"].values, edges_df_L["target"].values),
),
dtype=np.float64,
shape=(vertex_count_L, vertex_count_L),
)
CPU times: user 127 ms, sys: 43.9 ms, total: 171 ms
Wall time: 169 ms
Forward star in Cython
This code is adapted from the scipy.sparse source code coo_tocsr that can be found here on github, written in C++. Some important features are listed in the file header:
* Input: row and column indices *are not* assumed to be ordered
*
* Note: duplicate entries are carried over to the CSR represention
*
* Complexity: Linear. Specifically O(nnz(A) + max(n_row,n_col))
So this algorithm saves us from sorting the source node indices, which is rather expensive. Also, it handles duplicated entries, which might be a little bit confusing because the scipy.sparse.coo_array methods tocsr and tocsc do not. This means that duplicate entries are probably removed, by adding them together, downstream from this function in SciPy. Finally, the algorithm complexity is given. In our case, that would be:
where $|E|$ is the edge count, and $|V|$ the vertex count. A similar approach is described in [1].
In the following the cpdef function can be accessed from the Python level, while the cdef can only be seen in the Cython level.
%%cython --compile-args=-Ofast
# cython: language_level=3, boundscheck=False, wraparound=False, embedsignature=False, initializedcheck=False
import numpy as np
cimport numpy as cnp
cdef void coo_tocsr(
cnp.uint32_t [::1] Ai,
cnp.uint32_t [::1] Aj,
cnp.float64_t[::1] Ax,
cnp.uint32_t [::1] Bp,
cnp.uint32_t [::1] Bj,
cnp.float64_t[::1] Bx) nogil:
cdef:
ssize_t i, row, dest
ssize_t n_vert = <ssize_t>(Bp.shape[0] - 1)
ssize_t n_edge = <ssize_t>Bj.shape[0]
cnp.uint32_t temp, cumsum, last
for i in range(n_edge):
Bp[<ssize_t>Ai[i]] += 1
cumsum = 0
for i in range(n_vert):
temp = Bp[i]
Bp[i] = cumsum
cumsum += temp
Bp[n_vert] = <cnp.uint32_t>n_edge
for i in range(n_edge):
row = <ssize_t>Ai[i]
dest = <ssize_t>Bp[row]
Bj[dest] = Aj[i]
Bx[dest] = Ax[i]
Bp[row] += 1
last = 0
for i in range(n_vert + 1):
temp = Bp[i]
Bp[i] = last
last = temp
cpdef create_forward_star(edges_df, vertex_count, edge_count):
fs_indptr = np.zeros(vertex_count + 1, dtype=np.uint32) # make sure it is filled with zeros
fs_indices = np.empty(edge_count, dtype=np.uint32)
fs_data = np.empty(edge_count, dtype=np.float64)
coo_tocsr(
edges_df["source"].values,
edges_df["target"].values,
edges_df["weight"].values,
fs_indptr,
fs_indices,
fs_data)
return fs_indptr, fs_indices, fs_data
Here are a few remarks about this piece of code:
- a few compiler directives are invoked to speed up the code:
language_level,boundscheck,wraparound,embedsignature,initializedcheck - typed memoryviews are used to directly access NumPy arrays:
Typed memoryviews allow efficient access to memory buffers, such as those underlying NumPy array
This is why NumPy arrays are created before calling the coo_tocsr function
- input NumPy arrays of the
coo_tocsrfunction are declared as contiguous with the[::1]notation - the GIL is released with
nogilfor thecoo_tocsrfunction - array indices are declared as
ssize_ttype
Now we create a function that loops over an array of node indices and go though each node’s outgoing edges:
%%cython --compile-args=-Ofast
# cython: language_level=3, boundscheck=False, wraparound=False, embedsignature=False, initializedcheck=False
cimport numpy as cnp
cpdef loop_over_forward_star(
cnp.uint32_t [::1] node_seq,
cnp.uint32_t [::1] fs_indptr,
cnp.uint32_t [::1] fs_indices,
cnp.float64_t[::1] fs_data,
cnp.int8_t print_edges
):
cdef:
ssize_t i
ssize_t n_vert = <ssize_t>(fs_indptr.shape[0] - 1)
cnp.uint32_t tail_vert_idx, head_vert_idx, indptr
cnp.float64_t edge_data
if print_edges:
for i in range(n_vert):
tail_vert_idx = node_seq[i]
for indptr in range(fs_indptr[<ssize_t>tail_vert_idx], fs_indptr[<ssize_t>(tail_vert_idx + 1)]):
head_vert_idx = fs_indices[<ssize_t>indptr]
edge_data = fs_data[<ssize_t>indptr]
print(f"({tail_vert_idx},{head_vert_idx}) : {edge_data}")
else:
with nogil:
for i in range(n_vert):
tail_vert_idx = node_seq[i]
for indptr in range(fs_indptr[<ssize_t>tail_vert_idx], fs_indptr[<ssize_t>(tail_vert_idx + 1)]):
head_vert_idx = fs_indices[<ssize_t>indptr]
edge_data = fs_data[<ssize_t>indptr]
This function has a print_edges boolean argument. If False, there is no print and the GIL is released.
Small network example
Let’s try this create_forward_star function with the small network:
%%time
fs_indptr_S, fs_indices_S, fs_data_S = create_forward_star(
edges_df_S, vertex_count_S, edge_count_S
)
CPU times: user 378 µs, sys: 132 µs, total: 510 µs
Wall time: 524 µs
fs_indptr_S
array([0, 2, 3, 3, 4], dtype=uint32)
fs_indices_S
array([1, 1, 3, 3], dtype=uint32)
fs_data_S
array([2., 1., 2., 3.])
Everything seems to be OK. Let’s loop over each node’s outgoing edges:
loop_over_forward_star(
np.arange(vertex_count_S, dtype=np.uint32),
fs_indptr_S,
fs_indices_S,
fs_data_S,
print_edges=True,
)
(0,1) : 2.0
(0,1) : 1.0
(1,3) : 2.0
(3,3) : 3.0
Larger network
Now we run the create_forward_star function on the larger network:
%%time
fs_indptr_L, fs_indices_L, fs_data_L = create_forward_star(
edges_df_L, vertex_count_L, edge_count_L
)
CPU times: user 253 ms, sys: 100 ms, total: 353 ms
Wall time: 351 ms
%%time
sp_csr_L = sp_coo_L.tocsr()
CPU times: user 496 ms, sys: 113 ms, total: 609 ms
Wall time: 610 ms
assert np.allclose(fs_indptr_L, sp_csr_L.indptr)
assert np.allclose(fs_indices_L, sp_csr_L.indices)
assert np.allclose(fs_data_L, sp_csr_L.data)
Cython loop over the forward star
We are going to use the random node sequence created earlier to go over all the nodes of the graph, and get the outgoing edge information, head vertex index and edge weight, for each node.
%%time
loop_over_forward_star(
node_randseq_L, fs_indptr_L, fs_indices_L, fs_data_L, print_edges=False
)
CPU times: user 46 µs, sys: 5 µs, total: 51 µs
Wall time: 65.3 µs
Quite fast!??
Reverse star in Cython
We do the same thing as with the forward star, but with the reverse star…
%%cython --compile-args=-Ofast
# cython: language_level=3, boundscheck=False, wraparound=False, embedsignature=False, cdivision=True, initializedcheck=False
import numpy as np
cimport numpy as cnp
cdef void coo_tocsc(
cnp.uint32_t [::1] Ai,
cnp.uint32_t [::1] Aj,
cnp.float64_t[::1] Ax,
cnp.uint32_t [::1] Bp,
cnp.uint32_t [::1] Bi,
cnp.float64_t[::1] Bx) nogil:
cdef:
ssize_t i, col, dest
ssize_t n_vert = <ssize_t>(Bp.shape[0] - 1)
ssize_t n_edge = <ssize_t>Bi.shape[0]
cnp.uint32_t temp, cumsum, last
for i in range(n_edge):
Bp[<ssize_t>Aj[i]] += 1
cumsum = 0
for i in range(n_vert):
temp = Bp[i]
Bp[i] = cumsum
cumsum += temp
Bp[<ssize_t>n_vert] = <cnp.uint32_t>n_edge
for i in range(n_edge):
col = <ssize_t>Aj[i]
dest = <ssize_t>Bp[col]
Bi[dest] = Ai[i]
Bx[dest] = Ax[i]
Bp[col] += 1
last = 0
for i in range(n_vert + 1):
temp = Bp[i]
Bp[i] = last
last = temp
cpdef create_reverse_star(edges_df, vertex_count, edge_count):
rs_indptr = np.zeros(vertex_count + 1, dtype=np.uint32) # make sure it is filled with zeros
rs_indices = np.empty(edge_count, dtype=np.uint32)
rs_data = np.empty(edge_count, dtype=np.float64)
coo_tocsc(
edges_df["source"].values,
edges_df["target"].values,
edges_df["weight"].values,
rs_indptr,
rs_indices,
rs_data)
return rs_indptr, rs_indices, rs_data
And here is a function that loops over a list of node indices and go though each node’s incoming edges:
%%cython --compile-args=-Ofast
# cython: language_level=3, boundscheck=False, wraparound=False, embedsignature=False, initializedcheck=False
cimport numpy as cnp
cpdef loop_over_reverse_star(
cnp.uint32_t [::1] node_seq,
cnp.uint32_t [::1] rs_indptr,
cnp.uint32_t [::1] rs_indices,
cnp.float64_t[::1] rs_data,
cnp.int8_t print_edges
):
cdef:
ssize_t i
ssize_t n_vert = <ssize_t>(rs_indptr.shape[0] - 1)
cnp.uint32_t tail_vert_idx, head_vert_idx, indptr
cnp.float64_t edge_data
if print_edges:
for i in range(n_vert):
head_vert_idx = node_seq[i]
for indptr in range(rs_indptr[<ssize_t>head_vert_idx], rs_indptr[<ssize_t>(head_vert_idx + 1)]):
tail_vert_idx = rs_indices[<ssize_t>indptr]
edge_data = rs_data[<ssize_t>indptr]
print(f"({tail_vert_idx},{head_vert_idx}) : {edge_data}")
else:
with nogil:
for i in range(n_vert):
head_vert_idx = node_seq[i]
for indptr in range(rs_indptr[<ssize_t>head_vert_idx], rs_indptr[<ssize_t>(head_vert_idx + 1)]):
tail_vert_idx = rs_indices[<ssize_t>indptr]
edge_data = rs_data[<ssize_t>indptr]
Small network example
%%time
rs_indptr_S, rs_indices_S, rs_data_S = create_reverse_star(
edges_df_S, vertex_count_S, edge_count_S
)
CPU times: user 186 µs, sys: 21 µs, total: 207 µs
Wall time: 219 µs
rs_indptr_S
array([0, 0, 2, 2, 4], dtype=uint32)
rs_indices_S
array([0, 0, 1, 3], dtype=uint32)
rs_data_S
array([2., 1., 2., 3.])
Again, everything seems to be OK. Let’s loop over each node’s incoming edges:
loop_over_reverse_star(
np.arange(vertex_count_S, dtype=np.uint32),
rs_indptr_S,
rs_indices_S,
rs_data_S,
print_edges=True,
)
(0,1) : 2.0
(0,1) : 1.0
(1,3) : 2.0
(3,3) : 3.0
Larger network
We run the create_reverse_star function on the larger network:
%%time
rs_indptr_L, rs_indices_L, rs_data_L = create_reverse_star(
edges_df_L, vertex_count_L, edge_count_L
)
CPU times: user 479 ms, sys: 105 ms, total: 585 ms
Wall time: 577 ms
%%time
sp_csc_L = sp_coo_L.tocsc()
CPU times: user 629 ms, sys: 75.7 ms, total: 705 ms
Wall time: 701 ms
assert np.allclose(rs_indptr_L, sp_csc_L.indptr)
assert np.allclose(rs_indices_L, sp_csc_L.indices)
assert np.allclose(rs_data_L, sp_csc_L.data)
Cython loop over the reverse star
This time, we are going to use the random node sequence created earlier to go through all the nodes of the graph, and get the incoming edge information, tail vertex index and edge weight, for each node.
%%time
loop_over_reverse_star(
node_randseq_L, rs_indptr_L, rs_indices_L, rs_data_L, print_edges=False
)
CPU times: user 54 µs, sys: 5 µs, total: 59 µs
Wall time: 74.1 µs
Forward star with a structured array
We now focus on the forward star Cython code. What if instead of having two distinct arrays of size $|E|$: fs_indices of type np.uint32 and fs_data of type np.float, we use a single array with size $(|E|, 2)$ and heterogeneous column types? This can be done using NumPy structured arrays. However, the tricky part is to access this structured array from Cython with a heterogoneaous dtype memoryview. Fortunately, there is a simple example in the Cython documentation that we used in the following.
%%cython --compile-args=-Ofast
# cython: language_level=3, boundscheck=False, wraparound=False, embedsignature=False, initializedcheck=False
import numpy as np
cimport numpy as cnp
CSR_EDGE_DTYPE = np.dtype([
('head', np.uint32),
('weight', np.float64)
])
cdef packed struct csr_edge_dtype_struct:
# The struct needs to be packed since by default numpy dtypes aren't aligned
cnp.uint32_t head
cnp.float64_t weight
cpdef coo_tocsr_struct(
cnp.uint32_t [::1] Ai,
cnp.uint32_t [::1] Aj,
cnp.float64_t[::1] Ax,
cnp.uint32_t [::1] Bp,
csr_edge_dtype_struct[::1] Bx):
cdef:
ssize_t i, row, dest
ssize_t n_vert = <ssize_t>(Bp.shape[0] - 1)
ssize_t n_edge = <ssize_t>Bx.shape[0]
cnp.uint32_t temp, cumsum, last
with nogil:
for i in range(n_edge):
Bp[<ssize_t>Ai[i]] += 1
cumsum = 0
for i in range(n_vert):
temp = Bp[i]
Bp[i] = cumsum
cumsum += temp
Bp[n_vert] = <cnp.uint32_t>n_edge
for i in range(n_edge):
row = <ssize_t>Ai[i]
dest = <ssize_t>Bp[row]
Bx[dest].head = Aj[i]
Bx[dest].weight = Ax[i]
Bp[row] += 1
last = 0
for i in range(n_vert + 1):
temp = Bp[i]
Bp[i] = last
last = temp
cpdef create_forward_star_struct(edges_df, vertex_count, edge_count):
fs_indptr = np.zeros(vertex_count + 1, dtype=np.uint32) # make sure it is filled with zeros
fs_edges = np.empty(edge_count, dtype=CSR_EDGE_DTYPE)
coo_tocsr_struct(
edges_df["source"].values,
edges_df["target"].values,
edges_df["weight"].values,
fs_indptr,
fs_edges)
return fs_indptr, fs_edges
%%time
fs_indptr_struct_S, fs_edges_struct_S = create_forward_star_struct(
edges_df_S, vertex_count_S, edge_count_S
)
CPU times: user 84 µs, sys: 8 µs, total: 92 µs
Wall time: 97 µs
As explained in [2] by Jake VanderPlas, the data is now arranged together in one convenient block of memory. We can now access each edge attribute with a dictionary key:
fs_edges[i]["head"]fs_edges[i]["weight"]
fs_edges_struct_S
array([(1, 2.), (1, 1.), (3, 2.), (3, 3.)],
dtype=[('head', '<u4'), ('weight', '<f8')])
Here is the code to loop over each node’s outgoing edges:
for tail_vert_idx in range(vertex_count_S):
for indptr in range(
fs_indptr_struct_S[tail_vert_idx], fs_indptr_struct_S[tail_vert_idx + 1]
):
head_vert_idx = fs_edges_struct_S[indptr]["head"]
edge_data = fs_edges_struct_S[indptr]["weight"]
print(f"({tail_vert_idx},{head_vert_idx}) : {edge_data}")
(0,1) : 2.0
(0,1) : 1.0
(1,3) : 2.0
(3,3) : 3.0
Adjacency Lists
Finally, we are going to use another data representation: the adjacency list. For each node, we store a list of outgoing edges with head node vertices and edge weights. These lists have a variable size depending on the number of outgoing edges. An adjacency list is usually implemented with a doubly linked list, which requires a lot of memory. Here we are going to implement it with pointers and structs, which is sometimes referred to as adjacency vectors.
The issue is that this data structure cannot persist in the Python level, it can only live within the Cython level. This is why we wrote a global function loop_AL to perfomer the adjacency initialization, the loop over each node’s outgoing edges, and the memory release.
The memory allocation step for adjacency list approach is rather slow, as compare to the forward/reverse star data structure. As described in [3], the adjacency vector and the forward/reverse star representations have similar efficiency when used in graph algorithms, but the setup time and used memory is larger for the adjacency list approach.
Note that we use the CSR representation of the graph as input to create the adjacency lists.
%%cython --compile-args=-Ofast
# cython: language_level=3, boundscheck=False, wraparound=False, embedsignature=False, initializedcheck=False
from time import perf_counter
cimport numpy as cnp
from libc.stdlib cimport free, malloc
cdef struct AdjacencyList:
ssize_t size
cnp.uint32_t* vertices
cnp.float64_t* weights
cdef struct AdjacencyLists:
ssize_t vertex_count
AdjacencyList* neighbors
cdef void init_AL(AdjacencyLists* adj, ssize_t vertex_count):
adj.vertex_count = vertex_count
adj.neighbors = <AdjacencyList*> malloc(vertex_count * sizeof(AdjacencyList))
cdef void create_AL(
AdjacencyLists* adj,
cnp.uint32_t [::1] csr_indptr,
cnp.uint32_t [::1] csr_indices,
cnp.float64_t[::1] csr_data) nogil:
cdef:
ssize_t i, tail_vert_idx, size, ptr
for tail_vert_idx in range(adj.vertex_count):
size = <ssize_t>(csr_indptr[tail_vert_idx + 1] - csr_indptr[tail_vert_idx])
adj.neighbors[tail_vert_idx].size = size
adj.neighbors[tail_vert_idx].vertices = <cnp.uint32_t*> malloc(size * sizeof(cnp.uint32_t))
adj.neighbors[tail_vert_idx].weights = <cnp.float64_t*> malloc(size * sizeof(cnp.float64_t))
for i in range(size):
ptr = <ssize_t>(csr_indptr[tail_vert_idx]) + i
adj.neighbors[tail_vert_idx].vertices[i] = csr_indices[ptr]
adj.neighbors[tail_vert_idx].weights[i] = csr_data[ptr]
cdef void loop_AL_inner(
cnp.uint32_t [::1] node_seq,
AdjacencyLists* adj) nogil:
cdef:
ssize_t i, j, tail_vert_idx
cnp.uint32_t head_vert_idx
cnp.float64_t edge_weight
ssize_t n_vert = <ssize_t>(node_seq.shape[0] - 1)
for i in range(n_vert):
tail_vert_idx = <ssize_t>node_seq[i]
for i in range(adj.neighbors[tail_vert_idx].size):
head_vert_idx = adj.neighbors[tail_vert_idx].vertices[i]
edge_weight = adj.neighbors[tail_vert_idx].weights[i]
cdef void free_AL(AdjacencyLists* adj) nogil:
cdef:
ssize_t tail_vert_idx
for tail_vert_idx in range(adj.vertex_count):
free(adj.neighbors[tail_vert_idx].vertices)
free(adj.neighbors[tail_vert_idx].weights)
free(adj.neighbors)
cpdef void loop_AL(
cnp.uint32_t [::1] node_seq,
cnp.uint32_t[::1] csr_indptr,
cnp.uint32_t[::1] csr_indices,
cnp.float64_t[::1] csr_data,
int vertex_count):
cdef:
AdjacencyLists adj
start = perf_counter()
init_AL(&adj, vertex_count)
end = perf_counter()
elapsed_time = end - start
print(f"AL init - Elapsed time: {elapsed_time:12.8f} s")
start = perf_counter()
create_AL(&adj, csr_indptr, csr_indices, csr_data)
end = perf_counter()
elapsed_time = end - start
print(f"AL create - Elapsed time: {elapsed_time:12.8f} s")
start = perf_counter()
loop_AL_inner(node_seq, &adj)
end = perf_counter()
elapsed_time = end - start
print(f"AL loop - Elapsed time: {elapsed_time:12.8f} s")
start = perf_counter()
free_AL(&adj)
end = perf_counter()
elapsed_time = end - start
print(f"AL free - Elapsed time: {elapsed_time:12.8f} s")
loop_AL(node_randseq_L, fs_indptr_L, fs_indices_L, fs_data_L, vertex_count_L)
AL init - Elapsed time: 0.00004646 s
AL create - Elapsed time: 3.23047482 s
AL loop - Elapsed time: 0.00000054 s
AL free - Elapsed time: 0.97683480 s
Conclusion
Forward and reverse stars appear to be the most efficient approach in our case, where:
- we do not want to edit the network topology,
- the networks are relatively sparse and small, as compared to some social networks for example.
As stated in [1]:
Compressed sparse row is typically the best format for sparse graphs, provided that new edges aren’t added and relatively few edges are deleted. CSR is compact, avoiding the memory waste of adjacency lists and vectors, and its memory footprint can be calculated directly from V and E. CSR is furthermore contiguous in memory, eliminating the time overhead of pointer chasing. It’s easy to persist CSR in memory-mapped files, and CSR is convenient once you become accustomed to it. The two-pass construction approach implemented above is asymptotically faster than sorting an edge list.
References
[1] Terence Kelly, Programming Workbench: Compressed Sparse Row Format for Representing Graphs. Usenix Mag. 45(4) [2020]. pdf
[2] Jake VanderPlas, Python data science handbook : essential tools for working with data. Sebastopol, CA: O’Reilly Media Inc. [2016] ISBN: 978-1491912058 online
[3] Mahammad Valiyev, Graph Storage : How good is CSR really? [2017] pdf